Abstract
Mesenchymal stem-cell based therapies have been proposed as novel treatments for intervertebral disc degeneration, a prevalent and disabling condition associated with back pain. The development of these treatment strategies, however, has been hindered by the incomplete understanding of the human nucleus pulposus phenotype and by an inaccurate interpretation and translation of animal to human research. This review summarises recent work characterising the nucleus pulposus phenotype in different animal models and in humans and integrates their findings with the anatomical and physiological differences between these species. Understanding this phenotype is paramount to guarantee that implanted cells restore the native functions of the intervertebral disc.
Cite this article: Bone Joint Res 2013;2:169–78.
Introduction
Back pain is an almost universal symptom, with its prevalence by lifetime, month and time-point being 84%,1 23.2% and 11.9%,2 respectively. The economic impact of managing this condition and compensating for its associated losses and disabilities accounts for £12 billion annually in the United Kingdom3 and $85.9 billion in the USA.4
Although mechanical loading has been frequently been implicated in the pathogenesis of back pain,5-7 it is now known that genetic predisposition may account for a significant proportion of back pain associated clinical conditions.1,8,9 Significantly, MRI has shown that around 40% of patients with back pain have associated degeneration of the intervertebral disc (IVD),8,10 with a causal relationship between both having been identified.11-13 IVD degeneration, as described by Kirkaldy-Willis et al14 in their seminal work in 1978, triggers a cascade of events that lead to most of the degenerative spinal conditions treated in current clinical practice (disc bulging and herniation, spinal stenosis without spondylolisthesis, spinal stenosis with degenerative spondylolisthesis and degenerative scoliosis).14,15 Pain arising from disc degeneration may be caused by inflammatory responses triggered by degenerated disc tissue herniating into the spinal canal,16 nerve ingrowth into the IVD itself following annular and endplate ruptures,13,17 or due to altered spine biomechanics.18
The IVD is responsible for shock absorption and mobility of the spinal unit.19,20 It is composed of a central nucleus pulposus (NP) and a peripheral annulus fibrosus (AF), and is separated from the vertebral bodies by two cartilaginous endplates. The NP forms the gelatinous inner core of the IVD. It comprises large quantities of the proteoglycan aggrecan within an irregular mesh of type II collagen fibres. The AF is subdivided into outer AF, which is formed by distinct lamellae, composed of type I collagen fibres oriented obliquely between each lamellae,21,22 and a less fibrous and less organised inner AF, characterised by a transition to type II collagen and increased proteoglycan content.23 This architecture enables the AF to constrain the hydrostatic pressures generated within the NP upon compression, facilitating mobility between the spinal segments.24-27 The endplates are hyaline cartilage structures that form the interface between the IVD and the upper and lower platforms of the vertebral bodies. They protect the NP and AF inferiorly and superiorly and allow diffusion of oxygen and nutrients to the NP. Their structure is formed by a network of type II collagen fibrils and proteoglycans. Adjacent to the vertebral body the endplate is richer in collagen, whereas adjacent to the NP it is richer in proteoglycans.28
The adult IVD is practically devoid of blood vessels, being the largest avascular structure in the human body, and, therefore, relies on diffusion through the endplate for nutrient and oxygen supply. There is consequently a gradient of nutrient and oxygen availability from the peripheral AF to the central NP, where nutrient and oxygen concentrations are the lowest. As cells in the NP have to survive in oxygen-deprived conditions, they rely mostly on glycolysis to produce energy. In glycolysis, glucose is metabolised into its waste product, lactic acid, which is removed from the NP by diffusion through the endplate.29
NP cell morphology varies with age and maturation and across different animal species. In humans, NP cells are large and vacuolated at birth, similar to notochordal cells from which they are derived, but a few years after birth the NP becomes populated by smaller, round and non-vacuolated cells, resembling articular cartilage (AC) chondrocytes – for this reason they have been termed chondrocyte-like cells.30 This change in cell morphology has led to a long lasting debate regarding the origin of NP cells. Some authors argue that notochordal cells die and are replaced by chondrocyte-like cells migrating from adjacent tissues,31-33 whereas others suggest that the original notochordal NP cell population differentiates into smaller chondrocyte-like cells.34-38 Significantly, these differences in cell morphology are not common to all animal species with some animals retaining cells with notochordal morphology during most or even their entire lifespan.19 These interspecies differences together with the morphological similarity between human adult NP cells and articular chondrocytes have frequently led to an erroneous interpretation of NP cells as chondrocytes, and to inaccurate interpretations of research derived from animal studies.
Treatments for disc degeneration and the need for cell-based therapies
There is no definitive cure for disc degeneration and current surgical treatments for disc degeneration-associated conditions (such as herniation, stenosis or deformity) rely on discectomy, spinal fusion and disc replacement. These treatments, however, are associated with important complications, such as degeneration of the spinal levels adjacent to the fused/replaced one39 and prosthesis-related complications, such as migration into the adjacent vertebral body, extrusion and failure.40 More importantly, these treatments address the disease symptoms and not disease itself, failing to repair or regenerate the IVD.
Recent progress in tissue engineering and regenerative medicine has increased the interest in developing a biological approach to this disease, through which cells alone or together with biomaterials would be implanted into the NP to both repopulate and to stimulate native cells to produce a healthier extracellular matrix. The promising clinical results obtained in the treatment of osteoarthritis using autologous chondrocyte implantation and matrix-induced autologous chondrocyte implantation, in which autologous articular chondrocytes are harvested from non-damaged AC areas, expanded ex vivo and re-implanted (with or without a scaffold) into damaged osteo-arthritic regions of joints,41,42 have led researchers to attempt a similar strategy in the IVD. With this aim, dog IVD cells expanded ex vivo and re-implanted into its discs were shown to integrate and produce a cartilaginous extracellular matrix43; these results have led to the initiation of the Euro Disc Randomized Trial, an ongoing human clinical trial.44 Implantation of autologous NP cells harvested from degenerate discs may not be ideal, however, as degenerate NP cells have an altered phenotype, with increased expression of senescence markers,45 increased expression of matrix catabolic enzymes46,47 and decreased expression of matrix components48,49 factors that would impair the ability of these cells to produce a healthy matrix.
A different approach would be to harvest cells from non-degenerate discs. However, the method currently used to harvest cells from healthy discs (disc needle puncture) has been shown to induce degeneration.50-52 Alternatively, Nomura et al53 have proposed transplanting allogenic NP tissue or cells. The use of such cells or tissue, however, would require a donor bank of healthy human samples, which would be difficult to obtain and could pose immune rejection problems.
Stem cells, particularly mesenchymal stem cells, have also been widely proposed as a source of cells in the treatment of disc degeneration. A PubMed search including the terms (‘intervertebral disc’ or ‘spinal degeneration’ or ‘disc degeneration’ or ‘degenerative disc disease’) AND (‘stem cells’ or ‘stem cell’ or ‘mesenchymal cell’ or ‘stromal cells’ or ‘MSC’) retrieved 261 papers, of which over 50% have been published in the last three years. Mesenchymal stem cells are mesoderm-derived adult stem cells, for which there is a growing body of evidence confirming that they can be differentiated to adult NP cells,37,54-62 suggesting they may be the ideal candidates for novel cell-based therapies for disc degeneration.
Assessing stem cell differentiation: the importance of phenotyping the NP
For cell-based therapies to be successful, it is fundamental that implanted cells have the correct phenotype to produce an appropriate functioning matrix in vivo. Stem cell fate or differentiation can be influenced by co-culture, growth factors and/ or biophysical conditions.63,64 However, in order to identify which differentiating factors should be used and to assess the differentiation (particularly the “end-stage” cell created), it is important to understand the NP cell phenotype and specific cell markers.
To date, most of the studies assessing differentiation of MSC to NP cells analyse differentiation and ‘end-stage’ cell phenotype using traditional chondrogenic genes, such as collagen type II alpha 1 (COL2A1), aggrecan (ACAN) and sex determining region Y (SRY)-box 9 (SOX9),55,56,65-67 markers that are known to be expressed by healthy adult human NP cells.49
However, while NP cells have some similarities with AC cells, these cells and the tissues in which they reside have considerable differences in terms of cell ontogeny, morphology, matrix composition and biomechanical behaviour (Table I),68-71 and despite being important in the tissue’s function, they have not been taken into account when designing and assessing cell-based therapies for disc degeneration. This is highlighted by the study by Gorensek et al72 in which elastic cartilage from a rabbit’s ear was transplanted into its IVD and shown to form a solid tissue resembling AC, rather than a hydrated-gelatinous tissue like the NP. More recently, the relevance of these differences to the tissue’s biology has led to a growing interest in identifying specific NP markers, characteristic of its phenotype. Several years ago the proteins hypoxia inducible factors 1 alpha and beta (HIF-1α and HIF-1β), glucose transporter 1 (GLUT-1), matrix metalloproteinase 2 (MMP-2) and vascular endothelial growth factor (VEGF) were shown to have higher expression in the rat NP in comparison with its adjacent AF and cartilaginous endplate and thus, were proposed as NP-specific markers.73,74 However, these molecules are associated with responses to hypoxia and glucose starvation and may correspond to an adaptation of NP cells to the unique metabolic conditions NP cells have to withstand75 rather than marking a distinct cellular phenotype.
Table I
Main differences between articular cartilage and nucleus pulposus
| Characteristics | Articular cartilage | Nucleus pulposus | |
|---|---|---|---|
| Cell ontogeny34,35 | Lateral plate mesoderm | Notochord (axial mesoderm) | |
| Cell morphology | Small and round | Variable between species. In humans, they are large and vacuolated at birth, becoming small and round with maturation/degeneration | |
| Extracellular matrix proteoglycans68 | Large aggregates (hyaluronic acid and central filaments), multiple monomers and large non-aggregated monomers | Short non-aggregated proteoglycan monomers and clusters of monomers without apparent central filaments | |
| Aggrecan/type II collagen ratio69 | 2/1 | 27/1 | |
| Collagen network70 | Rigid | Loose | |
| Biomechanical behaviour71 | Viscoelastic solid in response to shear transient and dynamic deformation | Fluid under transient and of a viscoelastic solid under dynamic deformation | |
| Mechanical loads experienced71 | Compressive loading | Compressive and shear loading |
Consequently, a more thorough characterisation of these cells was needed and, with recent advances in transcriptomic profiling, the NP phenotype has now been described in several species.
Analysing the NP phenotype: gene expression profiling
In molecular biology, gene expression profiling is the measurement of the activity of genes being expressed by a given cell in a specific moment. Extensive characterisation of these genes is permitted by the use of powerful technologies, such as microarrays, ribonucleic acid (RNA)-sequencing and chromatin immuno-precipitation sequencing. The wide availability of microarrays has provided disease-related research with valuable transcriptomic information on the interactions between cells and the environment in which they reside with this information being used to characterise disease states, predict disease progression and develop new therapies.76-78
In the IVD field, microarrays have been used to characterise and define the NP cell phenotype, particularly by highlighting differences between the NP and similar or neighbouring tissues, such as AC, AF and endplate. Due to the difficulty in obtaining human intervertebral disc tissue, particularly non-degenerate, several of these studies have used animal models. Furthermore, for safety reasons, experimental treatments must be tested in animals and, therefore, it is important to understand if disease mechanisms differ between species and if/how they can be translated from animal research to human therapy. Phenotyping studies have thus been conducted on frequently used animal models, such as the rat,79-81 but also animals whose IVD is thought to more closely resemble the human, such as the dog82 and the bovine.36 More recently, the human NP phenotype has also been assessed.37,83
Results from these studies have provided new insights into NP markers in different species, but also on fundamental aspects regarding the ontogeny, maturation and degeneration of NP cells. As such, these findings have important implications for the understanding of the pathogenesis and treatment of disc degeneration and will be the focus of the following sections of this review.
The rat nucleus pulposus
The rat is a frequently used model for human diseases and has been extensively used to understand disc degeneration.84-87 At birth, the rat NP is populated by large vacuolated notochordal cells and this population of cells is reported to be maintained during the first 12 months of life, after which it starts to be replaced by a smaller population of chondrocyte-like cells.88
Fujita et al79 have compared the transcriptome of the rat NP (eight-week-old) with that of two avascular tissues (AF and tendon), five mesenchymal tissues (skeletal muscle, skin, blood, bone and bone marrow) and two neurogenic tissues (spinal cord and brain). This comparison, however, failed to include the AC, which, as aforementioned, is the tissue that more closely resembles the NP. The authors identified cluster differentiation 24 (CD24), a cell adhesion glycoprotein expressed at the surface of B-lymphocytes and differentiating neuroblasts, as a cell surface marker with significantly higher expression in the NP than in the other tissues analysed. Additionally, CD24 immunopositivity was shown to be present in six out of seven human chordomas (a notochord-derived bone tumour found in the spine) but not in chondrosarcoma samples.79
In a more recent study, Tang et al81 compared the NP and AF gene expression and again identified CD24, but also brain abundant membrane attached signal protein (BASP1), N-Cadherin (N-Cad), neuropilin (NRP-1), CD155 and CD221 as specific rat NP markers. Interestingly, CD24 has also been shown to be expressed by NP cells from six- to 17-year-old patients undergoing scoliosis surgery, having been proposed as an immature human NP-marker.89
In another rat phenotyping study, Lee et al80 identified annexin A3 (ANXA3), glypican 3 (GPC3), keratin 19 (KRT19) and pleiotrophin (PTN) as highly differentially expressed NP markers genes (compared with the AF and AC), with KRT19 and GPC3 differential expression being confirmed at the protein level. CD24, which had been proposed by Fujita et al79 as a NP-specific cell surface marker was also found to be highly differentially expressed in the NP, but failed to meet the criteria of at least five-fold differential expression used in this study. GPC3, KRT19 and PTN are all involved in tissue development and differentiation, with KRT19 in particular being expressed by the human embryonic notochord90 and chordoma.91 Interestingly, KRT19 expression in the rat NP is not restricted to the notochordal cell-rich NP and has also been found in adult (two-year-old) rat NP, where large vacuolated notochordal cells were reported to be absent.80
Stressing the importance of distinguishing the NP from the AC, this study found that the gene procol2a1, which codes for COL2A1, the predominant collagen type in NP and AC, although being highly expressed in both tissues, had five times higher expression in the AC,80 supporting the more rigid nature of the AC compared with the more gelatinous NP.
Despite being a commonly used animal model for disc degeneration, findings from studies in the rat may not be directly translatable to human research, as there are considerable cellular and microenvironmental differences between the IVD in both species. Being small quadrupeds, the load applied to the rat spine is substantially smaller than the load applied to the human spine.92 Additionally, as the human IVD cross-sectional area is significantly larger than the rat’s, and since nutrient intake and waste product removal from the NP are made through diffusion from the endplate,29 it is possible that the human NP is subjected to much more stressful conditions (e.g. glucose starvation, hypoxia and acidic pH) than the rat NP. Most importantly, rats retain morphologically distinct notochordal cells during at least half of their lifespan, whereas such cells disappear from the human NP soon after birth. These differences may have a considerable impact on the NP phenotype and on the translation of these findings to humans.
The canine nucleus pulposus
Another animal frequently used to study disc degeneration, particularly in NP phenotyping studies, is the dog.93-96 Dogs are larger quadrupeds and have a larger IVD than rats. Additionally, similar to humans, certain breeds of dogs – such as the dachshund, the beagle, the bulldog and the basset hound (together named chondrodystrophoid dogs) – are reported to lose their notochordal cells soon after birth; these species then undergo spontaneous disc degeneration.93,97 These aspects make these dog species potentially more suitable as a model for human disc degeneration.
Sakai et al82 performed comparative microarray analysis between the NP and AF from 16- to 18-month-old beagles and further analysed the identified molecules at the gene and protein level in NP, AF and AC from the same animals. Among the identified markers, α-2-macroglobulin (A2M), keratin-18 (KRT18) and neural cell adhesion molecule (NCAM1) were identified as highly expressed in the NP compared to the AF and AC and desmocolin-2 (DSC2) compared with the AF. The high differential expression of A2M in the aggrecan-rich dog NP may account for its biological function – A2M has been shown to be a potent inhibitor of the aggrecanases ADAMTS-4 and -5,98 two aggrecanases implicated in NP matrix degradation.46,99 NCAM1 (CD56) and DSC2 are two cell surface markers involved in cell-cell adhesion and interaction but their specific function in the dog NP remains to be explained. Like the rat NP marker KRT19, KRT18 has also been identified in human notochordal cells during development90 and also in the human NP.100,101
Of relevance is also the fact that this study identified significant differences between the dog and the rat NP phenotype. From the previously reported rat NP markers (ANXA3, GPC3, KRT19, PTN and CD24) only GPC3 was detected in this study and, contrary to the rat, it showed higher expression in the AF than in the NP. These differences in gene expression between species are possibly a reflection of the differences in the different cellular and biochemical environment in the NP of these two species.
The bovine nucleus pulposus
The finding that there are significant variations between the NP phenotype of different species triggered the search for the NP phenotype in other species, which would more closely resemble the human NP. Bovine caudal discs have been widely used as a model to study the human IVD.51,65,102,103 The IVD and the endplate of both species have been reported to have similar diameters and thicknesses and, due to the cow tail’s musculature, to be submitted to similar biomechanical forces.92 In terms of cellular population, the adult bovine NP predominantly contains a large population of small chondrocyte-like cells, although some authors have described a coexistent small population of large vacuolated notochord cells in animals aged < 30 months.19,36
A comparative microarray analysis was performed on bovine NP, AF and AC,36 which identified and further validated KRT8, KRT18, KRT19, N-Cad, synaptosomal-associated protein 25 (SNAP25) and sclerostin domain containing 1 (SOSTDC1) as specific NP markers; BASP1, KRT8, KRT18, tenomodulin (TNMD), TNF alpha induced protein 6 (TNFAIP6), N-cad, forkhead box F1 (FOXF1), forkhead box F2 (FOXF2), aquaporin 1 (AQP1), SOSTDC1 and SNAP25 as specific IVD (NP and AF) markers; and integrin binding sialoprotein (IBSP) and fibulin 1 (FBLN1) as negative NP markers. Confirming the interspecies variations that had been suggested in the canine array,82 this study failed to identify any of the rat NP markers GPC3, ANXA3, VIM, COMP, PTN and MGP or canine NP markers A2M, ANXA4, DSC2, NCAM1 as bovine NP markers.36
In an attempt to address the transcriptional profile of markers related to the unique IVD microenvironment, Minogue et al36 investigated the expression of previously identified rat microenvironment-related NP markers (MMP-2, HIF1A, GLUT-1 and VEGF) in bovine tissues, having failed, however, to identify significant differences between their gene expression in the bovine NP and AC. Although these differences may again be related to significant interspecies variations, it is of important note that these markers were originally identified by changes in protein expression and, therefore, the lack of differences in the bovine transcriptome may also be due to post-translational regulation or protein degradation, altering protein half-life, which would not be reflected at the mRNA level.104,105
Interestingly, the only common genes to the bovine NP phenotype and other species were KRT18 (dog NP marker), KRT19 (rat NP marker), KRT8 (rat NP marker identified by Lee et al80 but not chosen for qRT-PCR validation) and N-Cad (rat NP marker).
The human nucleus pulposus
The aforementioned transcriptional profiling studies were the first to provide a comprehensive list of NP markers for each specific species. They highlighted, however, the profound interspecies variations in gene expression, which possibly reflect differences in tissue and cell organisation, IVD and spine size and biomechanics and different local physicochemical environment in each species. This emphasised even more the need for the identification of the human NP genetic signature.
Such a study was recently performed by Minogue et al.37 Using a stringent criteria of 20-fold differential expression between the human NP and AC, the authors identified and validated FOXF1, ovostatin 2 (OVOS2), haemoglobin beta chain (HBB), carbonic anhydrase XII (CA12) and paired box 1 (PAX1) as specific human NP markers (compared with AC), and growth differentiation factor 10 (GDF10), integrin binding sialoprotein (IBSP) and cytokine-like 1 (CYTL1) as specific human AC (NP-negative) markers.37
PAX1 is a transcription factor expressed by the developing sclerotome and involved in the Shh-dependent mesenchymal-epithelial transition seen during the migration of sclerotomal cells towards the midline.106 The role of FOXF1 in the intervertebral disc has been less studied but recent findings have shown an association between deletions in the FOXF1 gene and the VACTERL association, a non-random association of birth defects which includes spinal malformations and fusion of spinal vertebrae.107
The high differential expression of HBB and CA12 is possibly related to the adaptive mechanisms of the NP cells to the harsh niche in which they reside. Haemoglobin, an oxygen transport metalloprotein – usually found in erythrocytes – has recently been identified in neurons, macrophages, alveolar cells and mesangial kidney cells where it has been proposed to act by storing oxygen under hypoxic conditions108-110; its identification in the hypoxic NP may represent a similar homeostatic mechanism. As NP cells have to survive in oxygen-deprived conditions, they rely mostly on glycolysis to produce energy and produce lactic acid as an end product; the accumulation of lactic acid in the NP leads to an acidic environment.111,112 CA12 is an enzyme that catalyses the conversion of carbon dioxide and water to bicarbonate and protons and is important in maintaining acid-base equilibrium by transporting carbon dioxide from cells residing in acidic environments113; together with CA9, it has also been identified in the developing IVD.114 It is possible that its presence in the human NP serves to counteract the accumulation of acidic waste products in this tissue.
Again, these results demonstrate high variability between species. From the previously identified rodent NP markers, only KRT19 and GPC3 showed a similar trend in the human arrays (higher differential expression in NP compared with AC); from the identified canine markers, a similar trend was found for DSC12, KRT18 and NCAM1; from the previously identified bovine markers, FOXF1 and FOXF2 were the only genes with at least two-fold differential expression between the two human tissues. Among the negative markers previously identified, the bovine markers IBSP (80-fold differential expression between both human tissues) and FBLN1 (qRT-PCR data) were also validated as negative human NP markers.
More recently, Power et al83 used a similar strategy to identify the human NP phenotype, and particularly NP-specific genes containing transmembrane domains, in comparison with the AF and AC. The rational for this strategy was, besides adding to information about the human NP phenotype, to identify potential NP-specific cell surface markers, which could be used to target specific cells with nanoparticles and, thereby, systemically delivering potential therapies to the NP cells.83 However, it should be noted that for such a strategy to be accomplished, the NP would have to have sufficient blood inflow to receive those particles, and the identified markers should not be expressed by any other cell in the human body, therefore assuring that the nanoparticles would only target NP cells. Surprisingly, the array data showed high similarity between the NP and the AF transcriptome and failed to identify a differentially expressed transmembrane-containing domain gene between both tissues. However, the comparison between NP and AC identified a list of 28 potential candidates, and their validation by qRT-PCR identified CA12 as the most significantly differentially expressed gene between the three tissues,83 adding strength to the importance of this acid-base regulator in the highly acidic NP niche.
Gene profiling provides clues for NP ontogeny
Results from these studies provide valuable information about the cellular population and the gene markers to be used in IVD research. Importantly they highlight that many of the markers are not common to all species and that, although they can be used for research in a specific animal model, they may not be suitable for translation to the human IVD in health and disease. Interestingly, they also highlight a significant overlapping gene profile across species for some of the identified markers. Of note, the following genes were found to have at least a two-fold differential expression between NP and AC or AF cells in at least two species: KRT8 (murine and bovine), KRT18 (murine, canine and human), KRT19 (murine and human), FOXF1 (bovine and human), BASP1 (rat and bovine) and N-Cad (rat and bovine) and may thus represent a common gene profile signature across these species (Table II).
Table II
List of identified nucleus pulposus markers
| Rat* | Dog* | Bovine* | Human* |
|---|---|---|---|
| KRT19†‡ | A2M | KRT8†‡ | FOXF1† |
| GPC3 | KRT18†‡ | KRT18†‡ | OVOS2 |
| ANXA3 | NCAM1 | KRT19†‡ | HBB |
| PTN | DSC2 | N-Cad† | CA12 |
| CD24 | SNAP25 | PAX1 | |
| KRT8†‡ | SOSTDC1 | KRT18†‡ | |
| N-Cad† | FOXF1† | KRT19†‡ | |
| FOXF2 | |||
-
* KRT, keratin; GPC, glypican; ANXA3, annexin a3; PTN, pleiotrophin; CD24, cluster differentiation 24; N-Cad, N-Cadherin; A2M, α-2-macroglobulin; NCAM, neural cell adhesion molecule; DSC, desmocolin; SNAP, synaptosomal-associated protein; SOSTDC1, sclerostin domain containing protein 1; FOXF1, forkhead box F1; OVOS, ovostatin; HBB, haemoglobin beta chain; CA12, carbonic anhydrase XII; PAX1, paired box gene 1 † indicates genes that have been identified in at least two different species ‡ indicates genes that indicate a notochordal ontogeny to the nucleus pulposus cells
Keratins comprise the largest subfamily of intermediate filaments proteins, proving structural support to the nucleus and tensile strength to the cell, but are also involved in more dynamic processes such as osmolarity regulation, protein synthesis, mitosis, cell movement and differentiation.115 Although keratin expression is traditionally indicative of an epithelial phenotype, its expression has been reported in mesoderm-derived cells, such as cardiomyocytes,116 fibroblasts117 and the human foetal notochord.90
Importantly, keratin expression in the NP provides clues to the ontogeny of the NP cells, which has been a subject of a long lasting debate. This controversy has been clarified by recent fate-mapping studies in mice where the embryonic notochord was shown to give rise to all the cells populating the adult mice NP.34,35 However, it was still not clear why the NP of some species lacks cells with a notochordal morphology. In order to further investigate the expression of keratin and also brachyury (also known to be expressed by notochordal cells118,119) in animals where notochordal cells are either absent or present in small numbers, we have isolated two subpopulations of bovine NP cells – large vacuolated and small chondrocyte-like (Fig. 1). Analysis of these two cell populations showed that, although these notochordal markers were highly expressed in larger cells, they were still very highly expressed in small chondrocyte-like cells, and that large cells share the expression of the chondrogenic markers (SOX9, COL2A1 and ACAN) with small NP cells (Fig. 2). This, together with the fact that keratins are expressed in the NP of all the animal species assessed to date, independently of the resident NP cell morphology, suggests that large vacuolated and small chondrocyte-like cells may all share a same notochordal lineage and that, contrary to what has been postulated, notochordal cells do not disappear but differentiate to smaller non-vacuolated cells. This supports previous findings of rabbit notochordal cells differentiating to chondrocyte-like cells as a response to injury.33
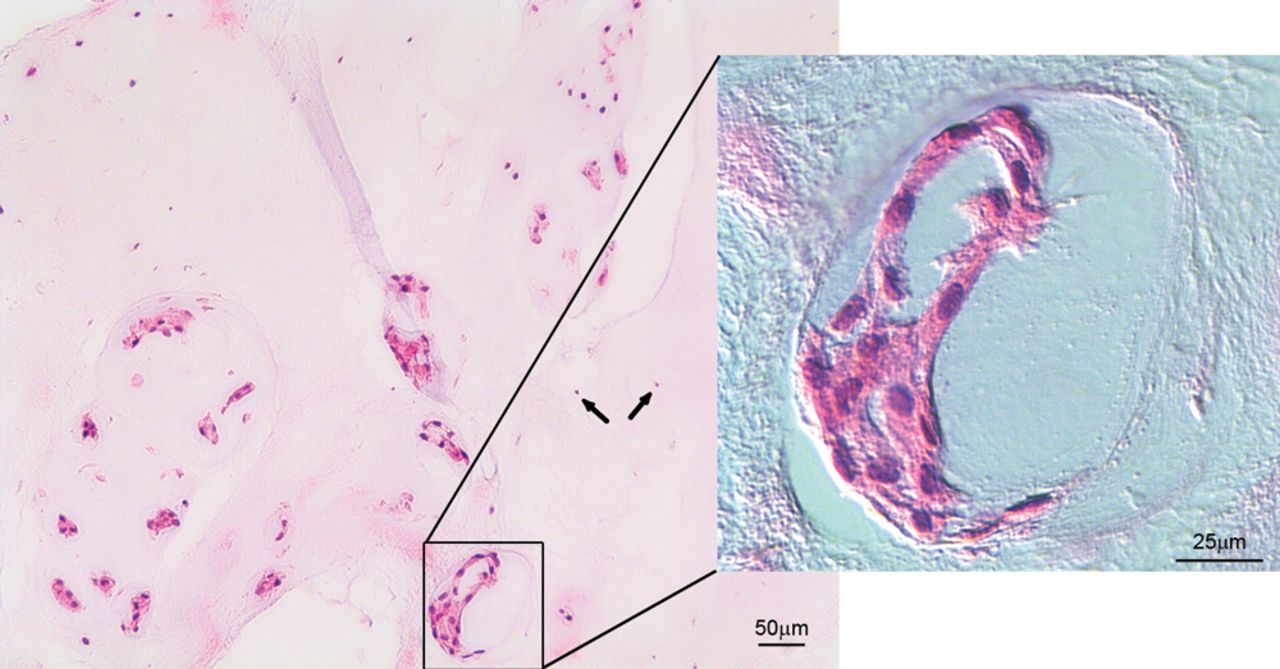
Fig. 1
Haematoxylin and eosin staining of bovine nucleus pulposus tissue, showing small chondrocyte-like cells (arrows) coexist with large vacuolated notochordal cells (magnified area).
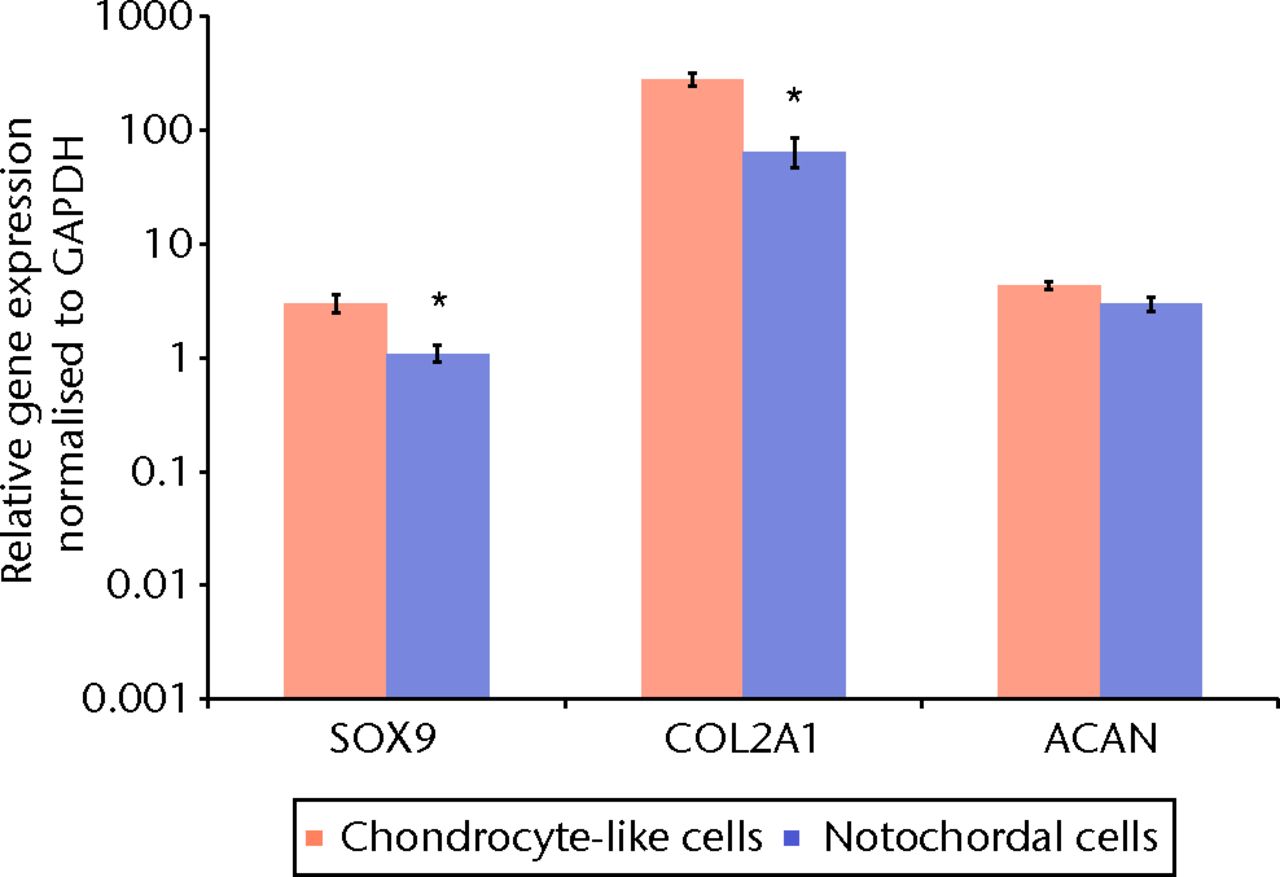
Fig. 2
Bar chart showing the quantitative real-time polymerase chain reaction for chondrogenic marker genes in separated bovine small chondrocyte-like and notochordal cells. The mean relative gene expression for the chondrogenic marker genes sex-determining region Y (SRY)-box 9 (SOX9), type II collagen (Col2A1) and aggrecan (ACAN) was normalised for the housekeeping gene glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and plotted on a log scale (* p < 0.05). Error bars denote the standard error of the mean.
Gene expression variations with degeneration
The identification of the novel NP markers has given not just a more comprehensive gene signature of the NP, but also triggered an interest as to whether this gene expression varies with ageing and disc degeneration.
Using the spontaneous dog model of disc degeneration (chondrodystrophic dogs), Smolders et al120 recently identified a down-regulation of genes involved in Wnt signalling and caveolin-1 occurring during the transition from a notochordal-rich to a chondrocyte-like-rich NP seen during early disc degeneration in these animals. The authors hypothesised that regulation of the Wnt pathway and of caveolin-1 expression in the IVD may be a therapeutic target for treatment of disc degeneration.120
In humans, a comparison between normal and degenerate NP samples has shown that with degeneration there is an alteration of the NP cell phenotype, with down-regulation of the expression of NP markers SNAP25, KRT-8, KRT-18 and N-Cad.36 This change has been confirmed by Weiler et al,101 who showed decreased immunopositivity for keratins and also galectin-3 with degeneration, although failing to distinguish between KRT8 and KRT19 immunopositivity. The decreased levels of SNAP25, KRT-8, KRT-18 and N-Cad with degeneration suggests that these may be markers of a healthier phenotype that are lost with degeneration. Conversely, KRT-19 and brachyury expression has been shown to remain unchanged with degeneration,36,121 which could then represent a stable NP signature throughout degeneration. Immunopositivity for CA12 was found to be high in the human immature NP, lower in the adult non-degenerate NP and higher with increasing degrees of degeneration, with its higher expression being proposed to correlate with the high metabolic developmental processes and as a response to injury.83
Interestingly, the AF marker FBLN1 was found to be significantly up-regulated in the NP of degenerate discs,36 which is in agreement with recent findings of AF cell migration to the NP during degeneration.122
These data support previous evidence that with human NP maturation, ageing and degeneration, the initial population of large, vacuolated notochordal cells does not disappear, but instead differentiates to a population of smaller chondrocyte-like cells; these morphologically distinct cells display some differences in their gene expression profile, but maintain the expression of genes, such as KRT19 and brachyury. This concept is supported by the findings of Yang et al,38 where the onset of degeneration was shown to coincide with differentiation of mouse notochordal to chondrocyte-like cells; however, the expression of putative NP or notochordal marker genes was not determined in this study.
Future perspectives
Transcriptional profiling of the NP has provided invaluable information that can help to establish the NP phenotype in various species. Researchers in the IVD field have now a library of specific genes that can be used to assess the NP phenotype, some of which are common to different animal species.
Additionally, the bovine and human studies have re-ignited a debate that had been ongoing for decades concerning the fate of notochordal cells in the NP of these species. Evidence now supports the premise that these cells do not disappear but rather differentiate to chondrocyte-like cells. Interestingly, some of the markers reflecting the notochordal ontogeny are also those that are common across species (KRT8, KRT18, KRT19 and brachyury). Conversely, other NP markers, such as HIF-1A, GLUT-1, HBB and CA12, may thus reflect responses to the microenvironmental conditions to which the NP cell is exposed to in each species.
As is norm in research, every scientific advance unveils undiscovered fields that warrant future research. It is yet to be clarified why in some animals NP cells maintain a notochordal morphology throughout life, whereas in others they differentiate to morphologically distinct cells. Additionally, is it not clear how homogeneous the small chondrocyte-like population of adult NP cells is. A recent report suggests that the expression of notochordal markers is restricted to subpopulations of the adult NP123 and that the non-notochordal population of NP cells may have migrated from the AF to the NP in response to degenerative stimuli.122 If so, it can be hypothesised that the elongated fibroblastic AF cells adopt a round chondrocyte-like morphology in response to the loose collagen network and high proteoglycan content in the NP, as opposed to the more ordered, fibrous matrix found in the AF. If this holds true, it remains to be shown whether these cells display different metabolism, response to injury, catabolic and anabolic properties. The identification of specific AF markers and, hence its phenotype, would allow identification of AF-derived cells with an NP-like morphology within the NP.
In respect to the treatment of intervertebral disc degeneration, particularly a cell-based therapy, it is still not know which phenotype should be targeted (young notochordal or a more mature chondrocyte-like). Understanding this is fundamental to guarantee that implanted cells have a NP phenotype capable of producing an appropriately functioning hydrated tissue that may regenerate and restore the functions of the intervertebral disc.
1 Balagué F , MannionAF, PelliséF, CedraschiC. Non-specific low back pain. Lancet2011;379:482–491. Google Scholar
2 Hoy DG , BainC, WilliamsG, et al.A systematic review of the global prevalence of low back pain. Arthritis Rheum2012;64:2028–2037.CrossrefPubMed Google Scholar
3 Maniadakis N , GrayA. The economic burden of back pain in the UK. Pain2000;84:95–103.CrossrefPubMed Google Scholar
4 Martin BI , DeyoRA, MirzaSK, et al.Expenditures and health status among adults with back and neck problems. JAMA2008;299:656–664.CrossrefPubMed Google Scholar
5 Bakker EW , VerhagenAP, LucasC, et al.Daily spinal mechanical loading as a risk factor for acute non-specific low back pain: a case-control study using the 24-Hour Schedule. Eur Spine J2007;16:107–113.CrossrefPubMed Google Scholar
6 Magnusson ML , AleksievA, WilderDG, et al.European Spine Society: the AcroMed Prize for Spinal Research 1995: unexpected load and asymmetric posture as etiologic factors in low back pain. Eur Spine J1996;5:23–35. Google Scholar
7 Svensson HO , AnderssonGB. Low-back pain in 40- to 47-year-old men: work history and work environment factors. Spine (Phila Pa 1976)1983;8:272–276.CrossrefPubMed Google Scholar
8 Livshits G , PophamM, MalkinI, et al.Lumbar disc degeneration and genetic factors are the main risk factors for low back pain in women: the UK Twin Spine Study. Ann Rheum Dis2011;70:1740–1745.CrossrefPubMed Google Scholar
9 Patel AA , SpikerWR, DaubsM, BrodkeD, Cannon-AlbrightLA. Evidence for an inherited predisposition to lumbar disc disease. J Bone Joint Surg [Am]2011;93-A:225–229.CrossrefPubMed Google Scholar
10 Cheung KM , KarppinenJ, ChanD, et al.Prevalence and pattern of lumbar magnetic resonance imaging changes in a population study of one thousand forty-three individuals. Spine (Phila Pa 1976)2009;34:934–940.CrossrefPubMed Google Scholar
11 Bogduk N . The lumbar disc and low back pain. Neurosurg Clin N Am1991;2:791–806. Google Scholar
12 Freemont AJ . The cellular pathobiology of the degenerate intervertebral disc and discogenic back pain. Rheumatology (Oxford)2009;48:5–10.CrossrefPubMed Google Scholar
13 Freemont AJ , PeacockTE, GoupilleP, et al.Nerve ingrowth into diseased intervertebral disc in chronic back pain. Lancet1997;350:178–181.CrossrefPubMed Google Scholar
14 Kirkaldy-Willis WH , WedgeJH, Yong-HingK, ReillyJ. Pathology and pathogenesis of lumbar spondylosis and stenosis. Spine (Phila Pa 1976)1978;3:319–328.CrossrefPubMed Google Scholar
15 Williams FM , PophamM, SambrookPN, et al.Progression of lumbar disc degeneration over a decade: a heritability study. Ann Rheum Dis2011;70:1203–1207.CrossrefPubMed Google Scholar
16 Ahn SH , ChoYW, AhnMW, et al.mRNA expression of cytokines and chemokines in herniated lumbar intervertebral discs. Spine (Phila Pa 1976)2002;27:911–917.CrossrefPubMed Google Scholar
17 Friberg S , HirschC. Anatomical and clinical studies on lumbar disc degeneration. Acta Orthop Scand1949;19:222–242.PubMed Google Scholar
18 Battie MC , VidemanT, LevalahtiE, GillK, KaprioJ. Heritability of low back pain and the role of disc degeneration. Pain2007;131:272–280.CrossrefPubMed Google Scholar
19 Hunter CJ , MatyasJR, DuncanNA. The notochordal cell in the nucleus pulposus: a review in the context of tissue engineering. Tissue Eng2003;9:667–677.CrossrefPubMed Google Scholar
20 Urban JP , McMullinJF. Swelling pressure of the lumbar intervertebral discs: influence of age, spinal level, composition, and degeneration. Spine (Phila Pa 1976)1988;13:179–187.CrossrefPubMed Google Scholar
21 Cassidy JJ , HiltnerA, BaerE. Hierarchical structure of the intervertebral discConnect Tissue. Res1989;23:75–88. Google Scholar
22 Marchand F , AhmedAM. Investigation of the laminate structure of lumbar disc anulus fibrosus. Spine (Phila Pa 1976)1990;15:402–410.CrossrefPubMed Google Scholar
23 Humzah MD , SoamesRW. Human intervertebral disc: structure and function. Anat Rec1988;220:337–356.CrossrefPubMed Google Scholar
24 Guerin HL , ElliottDM. Quantifying the contributions of structure to annulus fibrosus mechanical function using a nonlinear, anisotropic, hyperelastic model. J Orthop Res2007;25:508–516.CrossrefPubMed Google Scholar
25 Heuer F , SchmidtH, WilkeHJ. Stepwise reduction of functional spinal structures increase disc bulge and surface strains. J Biomech2008;41:1953–1960.CrossrefPubMed Google Scholar
26 Johannessen W , CloydJM, O’ConnellGD, VresilovicEJ, ElliottDM. Trans-endplate nucleotomy increases deformation and creep response in axial loading. Ann Biomed Eng2006;34:687–696.CrossrefPubMed Google Scholar
27 O’Connell GD , JohannessenW, VresilovicEJ, ElliottDM. Human internal disc strains in axial compression measured noninvasively using magnetic resonance imaging. Spine (Phila Pa 1976)2007;32:2860–2868.CrossrefPubMed Google Scholar
28 Hamilton DJ , SeguinCA, WangJ, PilliarRM, KandelRA. Formation of a nucleus pulposus-cartilage endplate construct in vitro. Biomaterials2006;27:397–405.CrossrefPubMed Google Scholar
29 Bibby SR , UrbanJP. Effect of nutrient deprivation on the viability of intervertebral disc cells. Eur Spine J2004;13:695–701.CrossrefPubMed Google Scholar
30 Trout JJ , BuckwalterJA, MooreKC. Ultrastructure of the human intervertebral disc: II: Cells of the nucleus pulposus. Anat Rec1982;204:307–314. Google Scholar
31 Butler WF. Comparative anatomy and development of the mammalian disc. In: Ghosh P, ed. The biology of the intervertebral disc. Boca Raton: CRC Press, 1989:83–108. Google Scholar
32 Kim KW , HaKY, LeeJS, et al.Notochordal cells stimulate migration of cartilage end plate chondrocytes of the intervertebral disc in in vitro cell migration assays. Spine J2009;9:323–329.CrossrefPubMed Google Scholar
33 Kim KW , LimTH, KimJG, et al.The origin of chondrocytes in the nucleus pulposus and histologic findings associated with the transition of a notochordal nucleus pulposus to a fibrocartilaginous nucleus pulposus in intact rabbit intervertebral discs. Spine (Phila Pa 1976)2003;28:982–990.CrossrefPubMed Google Scholar
34 Choi KS , CohnMJ, HarfeBD. Identification of nucleus pulposus precursor cells and notochordal remnants in the mouse: implications for disk degeneration and chordoma formation. Dev Dyn2008;237:3953–3958.CrossrefPubMed Google Scholar
35 McCann MR , TamplinOJ, RossantJ, SeguinCA. Tracing notochord-derived cells using a Noto-cre mouse: implications for intervertebral disc development. Dis Model Mech2012;5:73–82.CrossrefPubMed Google Scholar
36 Minogue BM , RichardsonSM, ZeefLA, FreemontAJ, HoylandJA. Transcriptional profiling of bovine intervertebral disc cells: implications for identification of normal and degenerate human intervertebral disc cell phenotypes. Arthritis Res Ther2010;12:R22.CrossrefPubMed Google Scholar
37 Minogue BM , RichardsonSM, ZeefLA, FreemontAJ, HoylandJA. Characterization of the human nucleus pulposus cell phenotype and evaluation of novel marker gene expression to define adult stem cell differentiation. Arthritis Rheum2010;62:3695–3705.CrossrefPubMed Google Scholar
38 Yang F , LeungVY, LukKD, ChanD, CheungKM. Injury-induced sequential transformation of notochordal nucleus pulposus to chondrogenic and fibrocartilaginous phenotype in the mouse. J Pathol2009;218:113–121.CrossrefPubMed Google Scholar
39 Lund T , OxlandTR. Adjacent level disk disease: is it really a fusion disease?Orthop Clin North Am2011;42:529–541. Google Scholar
40 Errico TJ . Lumbar disc arthroplasty. Clin Orthop Relat Res2005;435:106–117. Google Scholar
41 Marlovits S , AldrianS, WondraschB, et al.Clinical and radiological outcomes 5 years after matrix-induced autologous chondrocyte implantation in patients with symptomatic, traumatic chondral defects. Am J Sports Med2012;40:2273–2280.CrossrefPubMed Google Scholar
42 Moradi B , SchonitE, NierhoffC, et al.First-Generation autologous chondrocyte implantation in patients with cartilage defects of the knee: 7 to 14 years’ clinical and magnetic resonance imaging follow-up evaluation. Arthroscopy2012;28:1851–1861. Google Scholar
43 Meisel HJ , SiodlaV, GaneyT, et al.Clinical experience in cell-based therapeutics: disc chondrocyte transplantation A treatment for degenerated or damaged intervertebral disc. Biomol Eng2007;24:5–21.CrossrefPubMed Google Scholar
44 Hohaus C , GaneyTM, MinkusY, MeiselHJ. Cell transplantation in lumbar spine disc degeneration disease. Eur Spine J2008;17(Suppl 4):492–503.CrossrefPubMed Google Scholar
45 Le Maitre CL , FreemontAJ, HoylandJA. Accelerated cellular senescence in degenerate intervertebral discs: a possible role in the pathogenesis of intervertebral disc degeneration. Arthritis Res Ther2007;9:R45.CrossrefPubMed Google Scholar
46 Le Maitre CL , FreemontAJ, HoylandJA. Localization of degradative enzymes and their inhibitors in the degenerate human intervertebral disc. J Pathol2004;204:47–54.CrossrefPubMed Google Scholar
47 Le Maitre CL , FreemontAJ, HoylandJA. The role of interleukin-1 in the pathogenesis of human intervertebral disc degeneration. Arthritis Res Ther2005;7:R732–R745.CrossrefPubMed Google Scholar
48 Pearce RH , GrimmerBJ, AdamsME. Degeneration and the chemical composition of the human lumbar intervertebral disc. J Orthop Res1987;5:198–205.CrossrefPubMed Google Scholar
49 Sive JI , BairdP, JeziorskM, et al.Expression of chondrocyte markers by cells of normal and degenerate intervertebral discs. Mol Pathol2002;55:91–97.CrossrefPubMed Google Scholar
50 Carragee EJ , DonAS, HurwitzEL, et al.2009 ISSLS Prize Winner: Does discography cause accelerated progression of degeneration changes in the lumbar disc: a ten-year matched cohort study. Spine (Phila Pa 1976)2009;34:2338–2345.CrossrefPubMed Google Scholar
51 Michalek AJ , BuckleyMR, BonassarLJ, CohenI, IatridisJC. The effects of needle puncture injury on microscale shear strain in the intervertebral disc annulus fibrosus. Spine J2010;10:1098–1105.CrossrefPubMed Google Scholar
52 Nassr A , LeeJY, BashirRS, et al.Does incorrect level needle localization during anterior cervical discectomy and fusion lead to accelerated disc degeneration?Spine (Phila Pa 1976)2009;34:189–192.CrossrefPubMed Google Scholar
53 Nomura T , MochidaJ, OkumaM, NishimuraK, SakabeK. Nucleus pulposus allograft retards intervertebral disc degeneration. Clin Orthop Relat Res2001;389:94–101.CrossrefPubMed Google Scholar
54 Helder MN , KnippenbergM, Klein-NulendJ, WuismanPI. Stem cells from adipose tissue allow challenging new concepts for regenerative medicine. Tissue Eng2007;13:1799–1808.CrossrefPubMed Google Scholar
55 Henriksson HB , SvanvikT, JonssonM, et al.Transplantation of human mesenchymal stems cells into intervertebral discs in a xenogeneic porcine model. Spine (Phila Pa 1976)2009;34:141–148.CrossrefPubMed Google Scholar
56 Richardson SM , HughesN, HuntJA, FreemontAJ, HoylandJA. Human mesenchymal stem cell differentiation to NP-like cells in chitosan-glycerophosphate hydrogels. Biomaterials2008;29:85–93.CrossrefPubMed Google Scholar
57 Richardson SM , WalkerRV, ParkerS, et al.Intervertebral disc cell-mediated mesenchymal stem cell differentiation. Stem Cells2006;24:707–716.CrossrefPubMed Google Scholar
58 Risbud MV , AlbertTJ, GuttapalliA, et al.Differentiation of mesenchymal stem cells towards a nucleus pulposus-like phenotype in vitro: implications for cell-based transplantation therapy. Spine (Phila Pa 1976)2004;29:2627–2632.CrossrefPubMed Google Scholar
59 Sakai D , MochidaJ, IwashinaT, et al.Differentiation of mesenchymal stem cells transplanted to a rabbit degenerative disc model: potential and limitations for stem cell therapy in disc regeneration. Spine (Phila Pa 1976)2005;30:2379–2387.CrossrefPubMed Google Scholar
60 Stoyanov JV , Gantenbein-RitterB, BertoloA, et al.Role of hypoxia and growth and differentiation factor-5 on differentiation of human mesenchymal stem cells towards intervertebral nucleus pulposus-like cells. Eur Cell Mater2011;21:533–547.CrossrefPubMed Google Scholar
61 Strassburg S , HodsonNW, HillPI, RichardsonSM, HoylandJA. Bi-directional exchange of membrane components occurs during co-culture of mesenchymal stem cells and nucleus pulposus cells. PLoS One2012;7:33739.CrossrefPubMed Google Scholar
62 Strassburg S , RichardsonSM, FreemontAJ, HoylandJA. Co-culture induces mesenchymal stem cell differentiation and modulation of the degenerate human nucleus pulposus cell phenotype. Regen Med2010;5:701–711.CrossrefPubMed Google Scholar
63 Lutolf MP , GilbertPM, BlauHM. Designing materials to direct stem-cell fate. Nature2009;462:433–441.CrossrefPubMed Google Scholar
64 Murry CE , KellerG. Differentiation of embryonic stem cells to clinically relevant populations: lessons from embryonic development. Cell2008;132:661–680.CrossrefPubMed Google Scholar
65 Le Maitre CL , BairdP, FreemontAJ, HoylandJA. An in vitro study investigating the survival and phenotype of mesenchymal stem cells following injection into nucleus pulposus tissue. Arthritis Res Ther2009;11:R20.CrossrefPubMed Google Scholar
66 Richardson SM , CurranJM, ChenR, et al.The differentiation of bone marrow mesenchymal stem cells into chondrocyte-like cells on poly-L-lactic acid (PLLA) scaffolds. Biomaterials2006;27:4069–4078.CrossrefPubMed Google Scholar
67 Tao F , LiF, LiG, PanF. Differentiation of mesenchymal stem cells into nucleus pulposus cells in vitro. J Huazhong Univ Sci Technolog Med Sci2008;28:156–158.CrossrefPubMed Google Scholar
68 Buckwalter JA , SmithKC, KazarienLE, RosenbergLC, UngarR. Articular cartilage and intervertebral disc proteoglycans differ in structure: an electron microscopic study. J Orthop Res1989;7:146–151.CrossrefPubMed Google Scholar
69 Mwale F , RoughleyP, AntoniouJ. Distinction between the extracellular matrix of the nucleus pulposus and hyaline cartilage: a requisite for tissue engineering of intervertebral disc. Eur Cell Mater2004;8:58–64.CrossrefPubMed Google Scholar
70 Vonk LA , KroezeRJ, DoulabiBZ, et al.Caprine articular, meniscus and intervertebral disc cartilage: an integral analysis of collagen network and chondrocytes. Matrix Biol2010;29:209–218.CrossrefPubMed Google Scholar
71 Iatridis JC , WeidenbaumM, SettonLA, MowVC. Is the nucleus pulposus a solid or a fluid?: mechanical behaviors of the nucleus pulposus of the human intervertebral disc. Spine (Phila Pa 1976)1996;21:1174–1184. Google Scholar
72 Gorensek M , JaksimovicC, Kregar-VelikonjaN, et al.Nucleus pulposus repair with cultured autologous elastic cartilage derived chondrocytes. Cell Mol Biol Lett2004;9:363–373.PubMed Google Scholar
73 Fujita N , ImaiJ, SuzukiT, et al.Vascular endothelial growth factor-A is a survival factor for nucleus pulposus cells in the intervertebral disc. Biochem Biophys Res Commun2008;372:367–372.CrossrefPubMed Google Scholar
74 Rajpurohit R , RisbudMV, DucheyneP, VresilovicEJ, ShapiroIM. Phenotypic characteristics of the nucleus pulposus: expression of hypoxia inducing factor-1, glucose transporter-1 and MMP-2. Cell Tissue Res2002;308:401–407.CrossrefPubMed Google Scholar
75 Risbud MV , GuttapalliA, StokesDG, et al.Nucleus pulposus cells express HIF-1 alpha under normoxic culture conditions: a metabolic adaptation to the intervertebral disc microenvironment. J Cell Biochem2006;98:152–159.CrossrefPubMed Google Scholar
76 Karlsson C , DehneT, LindahlA, et al.Genome-wide expression profiling reveals new candidate genes associated with osteoarthritis. Osteoarthritis Cartilage2010;18:581–592.CrossrefPubMed Google Scholar
77 Saito T , FukaiA, MabuchiA, et al.Transcriptional regulation of endochondral ossification by HIF-2alpha during skeletal growth and osteoarthritis development. Nat Med2010;16:678–686.CrossrefPubMed Google Scholar
78 Yang S , KimJ, RyuJH, et al.Hypoxia-inducible factor-2alpha is a catabolic regulator of osteoarthritic cartilage destruction. Nat Med2010;16:687–693.CrossrefPubMed Google Scholar
79 Fujita N , MiyamotoT, ImaiJ, et al.CD24 is expressed specifically in the nucleus pulposus of intervertebral discs. Biochem Biophys Res Commun2005;338:1890–1896.CrossrefPubMed Google Scholar
80 Lee CR , SakaiD, NakaiT, et al.A phenotypic comparison of intervertebral disc and articular cartilage cells in the rat. Eur Spine J2007;16:2174–2185.CrossrefPubMed Google Scholar
81 Tang X , JingL, ChenJ. Changes in the molecular phenotype of nucleus pulposus cells with intervertebral disc aging. PLoS One2012;7:52020.CrossrefPubMed Google Scholar
82 Sakai D , NakaiT, MochidaJ, AliniM, GradS. Differential phenotype of intervertebral disc cells: microarray and immunohistochemical analysis of canine nucleus pulposus and anulus fibrosus. Spine (Phila Pa 1976)2009;34:1448–1456.CrossrefPubMed Google Scholar
83 Power KA , GradS, RutgesJP, et al.Identification of cell surface-specific markers to target human nucleus pulposus cells: expression of carbonic anhydrase XII varies with age and degeneration. Arthritis Rheum2011;63:3876–3886.CrossrefPubMed Google Scholar
84 Cassidy JD , Yong-HingK, Kirkaldy-WillisWH, WilkinsonAA. A study of the effects of bipedism and upright posture on the lumbosacral spine and paravertebral muscles of the Wistar rat. Spine (Phila Pa 1976)1988;13:301–308.CrossrefPubMed Google Scholar
85 Moskowitz RW , ZivI, DenkoCW, et al.Spondylosis in sand rats: a model of intervertebral disc degeneration and hyperostosis. J Orthop Res1990;8:401–411.CrossrefPubMed Google Scholar
86 Latorre A , AlbaredaJ, CastiellaT, LasierraJM, SeralF. Experimental model of multidirectional disc hernia in rats. Int Orthop 1998;22:44–48.CrossrefPubMed Google Scholar
87 Machida M , SaitoM, DuboussetJ, et al.Pathological mechanism of idiopathic scoliosis: experimental scoliosis in pinealectomized rats. Eur Spine J2005;14:843–848.CrossrefPubMed Google Scholar
88 Stevens JW , KurrigerGL, CarterAS, MaynardJA. CD44 expression in the developing and growing rat intervertebral disc. Dev Dyn2000;219:381–390.CrossrefPubMed Google Scholar
89 Tang X, Jing L, Setton L, et al. Identifying the molecular phenotype of cells in the human intervertebral disc reveals the existence of a unique notochordal-like cell population. Procs Orthopaedic Research Society Annual Meeting 2013, San Antonio, Texas. Google Scholar
90 Gotz W , KasperM, FischerG, HerkenR. Intermediate filament typing of the human embryonic and fetal notochord. Cell Tissue Res1995;280:455–462.CrossrefPubMed Google Scholar
91 Gottschalk D , FehnM, PattS, et al.Matrix gene expression analysis and cellular phenotyping in chordoma reveals focal differentiation pattern of neoplastic cells mimicking nucleus pulposus development. Am J Pathol2001;158:1571–1578.CrossrefPubMed Google Scholar
92 Alini M , EisensteinSM, ItoK, et al.Are animal models useful for studying human disc disorders/degeneration?Eur Spine J2008;17:2–19.CrossrefPubMed Google Scholar
93 Bray JP , BurbidgeHM. The canine intervertebral disk: Part two: degenerative changes--nonchondrodystrophoid versus chondrodystrophoid disks. J Am Anim Hosp Assoc1998;34:135–144. Google Scholar
94 Cole TC , BurkhardtD, GhoshP, RyanM, TaylorT. Effects of spinal fusion on the proteoglycans of the canine intervertebral disc. J Orthop Res1985;3:277–291.CrossrefPubMed Google Scholar
95 Olsewski JM , SchendelMJ, WallaceLJ, OgilvieJW, GundryCR. Magnetic resonance imaging and biological changes in injured intervertebral discs under normal and increased mechanical demands. Spine (Phila Pa 1976)1996;21:1945–1951.CrossrefPubMed Google Scholar
96 Puustjarvi K , LammiM, KivirantaI, HelminenHJ, TammiM. Proteoglycan synthesis in canine intervertebral discs after long-distance running training. J Orthop Res1993;11:738–746.CrossrefPubMed Google Scholar
97 Bray JP , BurbidgeHM. The canine intervertebral disk: part one: structure and function. J Am Anim Hosp Assoc1998;34:55–63.CrossrefPubMed Google Scholar
98 Tortorella MD , ArnerEC, HillsR, et al.Alpha2-macroglobulin is a novel substrate for ADAMTS-4 and ADAMTS-5 and represents an endogenous inhibitor of these enzymes. J Biol Chem2004;279:17554–17561.CrossrefPubMed Google Scholar
99 Pockert AJ , RichardsonSM, Le MaitreCL, et al.Modified expression of the ADAMTS enzymes and tissue inhibitor of metalloproteinases 3 during human intervertebral disc degeneration. Arthritis Rheum2009;60:482–491.CrossrefPubMed Google Scholar
100 Stosiek P , KasperM, KarstenU. Expression of cytokeratin and vimentin in nucleus pulposus cells. Differentiation1988;39:78–81.CrossrefPubMed Google Scholar
101 Weiler C , NerlichAG, SchaafR, et al.Immunohistochemical identification of notochordal markers in cells in the aging human lumbar intervertebral disc. Eur Spine J2010;19:1761–1770.CrossrefPubMed Google Scholar
102 Mwale F , CiobanuI, GiannitsiosD, et al.Effect of oxygen levels on proteoglycan synthesis by intervertebral disc cells. Spine (Phila Pa 1976)2011;36:E131–E138.CrossrefPubMed Google Scholar
103 Walter BA , KoreckiCL, PurmessurD, et al.Complex loading affects intervertebral disc mechanics and biology. Osteoarthritis Cartilage2011;19:1011–1018.CrossrefPubMed Google Scholar
104 Ideker T , ThorssonV, RanishJA, et al.Integrated genomic and proteomic analyses of a systematically perturbed metabolic network. Science2001;292:929–934.CrossrefPubMed Google Scholar
105 Yin D , GriffinMJ, EthertonTD. Analysis of the signal pathways involved in the regulation of fatty acid synthase gene expression by insulin and somatotropin. J Anim Sci2001;79:1194–1200.CrossrefPubMed Google Scholar
106 McMahon JA , TakadaS, ZimmermanLB, et al.Noggin-mediated antagonism of BMP signaling is required for growth and patterning of the neural tube and somite. Genes Dev1998;12:1438–1452.CrossrefPubMed Google Scholar
107 Stankiewicz P , SenP, BhattSS, et al.Genomic and genic deletions of the FOX gene cluster on 16q24.1 and inactivating mutations of FOXF1 cause alveolar capillary dysplasia and other malformations. Am J Hum Genet2009;84:780–791.CrossrefPubMed Google Scholar
108 Liu L , ZengM, StamlerJS. Hemoglobin induction in mouse macrophages. Proc Natl Acad Sci U S A1999;96:6643–6647.CrossrefPubMed Google Scholar
109 Newton DA , RaoKM, DluhyRA, BaatzJE. Hemoglobin is expressed by alveolar epithelial cells. J Biol Chem2006;281:5668–5676.CrossrefPubMed Google Scholar
110 Nishi H , InagiR, KatoH, et al.Hemoglobin is expressed by mesangial cells and reduces oxidant stress. J Am Soc Nephrol2008;19:1500–1508.CrossrefPubMed Google Scholar
111 Benneker LM , HeiniPF, AliniM, AndersonSE, ItoK. 2004 Young Investigator Award Winner: vertebral endplate marrow contact channel occlusions and intervertebral disc degeneration. Spine (Phila Pa 1976)2005;30:167–173.CrossrefPubMed Google Scholar
112 Neidlinger-Wilke C , MietschA, RinklerC, et al.Interactions of environmental conditions and mechanical loads have influence on matrix turnover by nucleus pulposus cells. J Orthop Res2012;30:112–121.CrossrefPubMed Google Scholar
113 Chiche J , IlcK, LaferriereJ, et al.Hypoxia-inducible carbonic anhydrase IX and XII promote tumor cell growth by counteracting acidosis through the regulation of the intracellular pH. Cancer Res2009;69:358–368.CrossrefPubMed Google Scholar
114 Liao SY , LermanMI, StanbridgeEJ. Expression of transmembrane carbonic anhydrases, CAIX and CAXII, in human development. BMC Dev Biol2009;9:22.CrossrefPubMed Google Scholar
115 Vaidya MM , KanojiaD. Keratins: markers of cell differentiation or regulators of cell differentiation?J Biosci2007;32:629–634. Google Scholar
116 Kuruc N , FrankeWW. Transient coexpression of desmin and cytokeratins 8 and 18 in developing myocardial cells of some vertebrate species. Differentiation1988;38:177–193.CrossrefPubMed Google Scholar
117 von Koskull H , VirtanenI. Induction of cytokeratin expression in human mesenchymal cells. J Cell Physiol1987;133:321–329.CrossrefPubMed Google Scholar
118 Herrmann BG , KispertA. The T genes in embryogenesis. Trends Genet1994;10:280–286.CrossrefPubMed Google Scholar
119 Vujovic S , HendersonS, PresneauN, et al.Brachyury, a crucial regulator of notochordal development, is a novel biomarker for chordomas. J Pathol2006;209:157–165.CrossrefPubMed Google Scholar
120 Smolders LA , MeijBP, OnisD, et al.Gene expression profiling of early intervertebral disc degeneration reveals a down-regulation of canonical Wnt signaling and caveolin-1 expression: implications for development of regenerative strategies. Arthritis Res Ther2013;15:R23.CrossrefPubMed Google Scholar
121 Risbud MV , SchaerTP, ShapiroIM. Toward an understanding of the role of notochordal cells in the adult intervertebral disc: from discord to accord. Dev Dyn2010;239:2141–2148.CrossrefPubMed Google Scholar
122 Tanaka MS , SakaiD, HiyamaA, et al.Evidence of non-notochordal origin in chondrocyte-like cells of the nucleus pulposus appearing in early stage disc degeneration in the mouse model. Global Spine J2012;2:S2–08. Google Scholar
123 Gilson A , DregerM, UrbanJP. Differential expression level of cytokeratin 8 in cells of the bovine nucleus pulposus complicates the search for specific intervertebral disc cell markers. Arthritis Res Ther2010;12:R24.CrossrefPubMed Google Scholar
Funding statement:
R. Rodrigues-Pinto is supported by a grant from the Programme for Advanced Medical Education, sponsored by Fundação Calouste Gulbenkian, Fundação Champalimaud, Ministério da Saúde, Fundação para a Ciência e Tecnologia and Apifarma, Portugal.
Author contributions:
R. Rodrigues-Pinto: Manuscript draft preparation, Relevant literature search and review
S. M. Richardson: Formulated manuscript outline, Reviewed and edited draft versions including final manuscript draft
J. A. Hoyland: Formulated manuscript outline, Reviewed and edited draft manuscript including final manuscript
ICMJE Conflict of Interest:
None declared
©2013 The British Editorial Society of Bone & Joint Surgery. This is an open-access article distributed under the terms of the Creative Commons Attributions licence, which permits unrestricted use, distribution, and reproduction in any medium, but not for commercial gain, provided the original author and source are credited.










