Abstract
Objective
Excessive mechanical stress on synovial joints causes osteoarthritis (OA) and results in the production of prostaglandin E2 (PGE2), a key molecule in arthritis, by synovial fibroblasts. However, the relationship between arthritis-related molecules and mechanical stress is still unclear. The purpose of this study was to examine the synovial fibroblast response to cyclic mechanical stress using an in vitro osteoarthritis model.
Method
Human synovial fibroblasts were cultured on collagen scaffolds to produce three-dimensional constructs. A cyclic compressive loading of 40 kPa at 0.5 Hz was applied to the constructs, with or without the administration of a cyclooxygenase-2 (COX-2) selective inhibitor or dexamethasone, and then the concentrations of PGE2, interleukin-1β (IL-1β), tumour necrosis factor-α (TNF-α), IL-6, IL-8 and COX-2 were measured.
Results
The concentrations of PGE2, IL-6 and IL-8 in the loaded samples were significantly higher than those of unloaded samples; however, the concentrations of IL-1β and TNF-α were the same as the unloaded samples. After the administration of a COX-2 selective inhibitor, the increased concentration of PGE2 by cyclic compressive loading was impeded, but the concentrations of IL-6 and IL-8 remained high. With dexamethasone, upregulation of PGE2, IL-6 and IL-8 was suppressed.
Conclusion
These results could be useful in revealing the molecular mechanism of mechanical stress in vivo for a better understanding of the pathology and therapy of OA.
Cite this article: Bone Joint Res 2014;3:280–8.
Article focus
Analysis of molecular mechanism of osteoarthritis (OA) development
Analysis of mechanotransduction in OA development
Function of synovial fibroblast in OA development
Key messages
Cyclic compressive loading on a 3D cultured construct of human fibroblasts upregulated PGE2 via COX-2 production
Cyclic compressive loading upregulated interleukin-6 (IL-6) and IL-8 proteins
The expression of these molecules was upregulated without IL-1β and/or tumour necrosis factor (TNF)-α stimulation
Strengths and limitations
Strengths - our 3D culture system is close to intra-articular environment
Our system could be useful in revealing the molecular mechanism of mechanical stress
Limitation - the intracellular signal transductions of PGE2, IL-6 and IL-8 (mechanotransduction) have not been clarified
Introduction
Osteoarthritis (OA) is a common disease that causes joint pain, deformity and functional disability, and is increasingly prevalent in hundreds of millions of people worldwide.1 Congenital disorders, obesity, labour, sports, malalignment and joint instability may initiate processes leading to loss of cartilage. In addition, repeated excessive mechanical stress on the synovial joint, which is composed of cartilage and synovium, is considered to be a key factor in OA development. However, the molecular relationship between mechanical stress and OA development is still unclear.
OA involves a variable degree of synovitis, and these inflammations cause many symptoms including joint swelling and effusion in clinical situations.2-6 Synovial fibroblasts and macrophages, as well as chondrocytes, play an important role in OA development through synovitis, and synovial macrophages are considered to produce pro-inflammatory cytokines, such as interleukin-1β (IL-1β) and tumour necrosis factor-α (TNF-α).6-10 These cytokines stimulate synovial fibroblasts and chondrocytes to produce other cytokines, such as IL-6 and IL-8, and several enzymes, such as matrix metalloproteinases (MMPs) and aggrecanases (ADAMTSs). These enzymes sever type II collagen and proteoglycan, the principal components of the extracellular matrix of articular cartilage.11-15 In addition, prostaglandin E2 (PGE2) plays a significant role in OA by causing pain, inflammation, and cartilage degradation.16-18 Although PGE2 is known to be produced by synovial fibroblasts or chondrocytes in response to IL-1β and/or TNF-α, which is produced by synovial macrophages,19-21 the molecular mechanism of PGE2 production triggered by mechanical stress is still unclear.
We have developed a novel three-dimensional (3D) culture system using cyclic mechanical stress on synovial cells or chondrocytes for revealing the molecular mechanism of OA development resulting from mechanical stress. In particular, we have focused on synovial cells, which play an important role in OA development as mentioned above. In our previous study, we have shown that cyclic mechanical stress on 3D cultured constructs of human synovial fibroblasts upregulated mRNA levels of MMP1, MMP2, MMP3, MMP9, MMP13, ADAMTS4, and ADAMTS5 genes in a load-dependent manner22-23 however, the induction of PGE2 as a result of mechanical stress has not been investigated. In the PGE2 synthesis pathway, cyclooxygenase-2 (COX-2) and microsomal prostaglandin E synthase-1 (mPGES-1) are key enzymes that metabolise arachidonic acid to PGE2.24,25 Nonsteroidal anti-inflammatory drugs (NSAIDs) and steroids, which downregulate PGE2 synthesis through inhibition of COX-2 activity, have been widely used in the treatment of OA.19
The purpose of this study was to examine the expression of PGE2 and the related cytokine expressions of IL-1β, TNF-α, IL-6 and IL-8 by cyclic compressive loading on 3D cultured constructs of human synovial fibroblasts and to clarify the effects of NSAIDs and steroids using our in vitro OAmodel.
Materials and Methods
Cell culture of primary human synovial fibroblasts
Human synovial membranes were obtained aseptically from eight patients aged from 17 to 34 years (three male, five female) who underwent arthroscopic knee surgery in accordance with a protocol approved by the Osaka University Institutional Ethical Committee. We followed the Helsinki Declaration and obtained written informed consent from all the patients involved in this study. The cell isolation protocol was essentially the same as the protocol used previously for the isolation of human synovial fibroblasts.22,26 In brief, synovial membrane specimens were rinsed with phosphate-buffered saline (PBS), minced meticulously and digested with 0.4% collagenase XI (Sigma-Aldrich, St. Louis, Missouri) for two hours at 37°C. After neutralisation of the collagenase with a growth medium containing high-glucose Dulbecco’s Modified Eagle’s Medium (HG-DMEM, Wako, Osaka, Japan) supplemented with 10% fetal bovine serum (FBS; HyClone, Logan, Utah) and 1% penicillin/streptomycin (Gibco BRL, Life Technologies Inc., Carlsbad, California), the cells were collected by centrifugation, washed with PBS, resuspended in a growth medium, and plated in culture dishes. For expansion, cells were cultured in the growth medium at 37°C in a humidified atmosphere of 5% CO2. The medium was replaced once a week. After ten to 14 days of primary culture, when the cells reached near confluence, they were washed twice with PBS, harvested by treatment with trypsin-EDTA (0.25% trypsin and 1 mM EDTA; Gibco BRL, Life Technologies Inc.), and replated at 1:3 dilution for the first subculture. Cell passages were continued in the same manner with 1:3 dilution when cultures reached near confluence. Cells at passages 3 to 7 were used in the present study.
Cell seeding on collagen scaffold and production of the 3D engineered construct
The primary cultured cells were harvested and seeded on collagen scaffolds to produce 3D constructs as previously described.22,23 In brief, the cultured cells (5 × 105/scaffold) were suspended in a growth medium and then mixed with an equal volume of 1% Atelocollagen gel (Koken, Tokyo, Japan) on ice to produce a cell suspension in 0.5% collagen solution. The cell suspension was incorporated into collagen scaffolds (Atelocollagen Sponge Mighty, Koken, Tokyo, Japan; 5 mm diameter, 3 mm thick) by centrifugation at 500 × g for five minutes. The collagen scaffold which we used has an interconnected pore size of 30 nm to 200 nm. The scaffolds were fabricated via the process of freeze-drying of 10% collagen gel and cross-linking to reinforce the mechanical property. This is similar to those of articular cartilage. The cell–scaffold constructs were then incubated at 37°C for gelation to produce 3D cell–scaffold constructs (Fig. 1a) were suspended in a growth medium and then mixed with an equal volume of 1% Atelocollagen gel (Koken, Tokyo, Japan) on ice to produce a cell suspension in 0.5% collagen solution. The cell suspension was incorporated into collagen scaffolds (Atelocollagen Sponge Mighty, Koken, Tokyo, Japan; 5 mm diameter, 3 mm thick) by centrifugation at 500 × g for five minutes. The collagen scaffold which we used has an interconnected pore size of 30 nm to 200 nm. The scaffolds were fabricated via the process of freeze-drying of 10% collagen gel and cross-linking to reinforce the mechanical property. This is similar to those of articular cartilage. The cell–scaffold constructs were then incubated at 37°C for gelation to produce 3D cell–scaffold constructs (Fig. 1a) on ice to produce a cell suspension in 0.5% collagen solution. The cell suspension was incorporated into collagen scaffolds (Atelocollagen Sponge Mighty, Koken, Tokyo, Japan; 5 mm diameter, 3 mm thick) by centrifugation at 500 × g for five minutes. The collagen scaffold which we used has an interconnected pore size of 30 nm to 200 nm. The scaffolds were fabricated via the process of freeze-drying of 10% collagen gel and cross-linking to reinforce the mechanical property. This is similar to those of articular cartilage. The cell–scaffold constructs were then incubated at 37°C for gelation to produce 3D cell–scaffold constructs (Fig. 1a) by centrifugation at 500 × g for five minutes. The collagen scaffold which we used has an interconnected pore size of 30 nm to 200 nm. The scaffolds were fabricated via the process of freeze-drying of 10% collagen gel and cross-linking to reinforce the mechanical property. This is similar to those of articular cartilage. The cell–scaffold constructs were then incubated at 37°C for gelation to produce 3D cell–scaffold constructs (Fig. 1a). The cells in the 3D construct were evenly embedded in the collagen scaffold, with no cell leakage and collagen breakage after cell seeding, as we have previously shown with histological evaluation.22 The constructs were maintained in a growth medium of HG-DMEM, with 10% FBS in free-swelling conditions at 37°C and in 5% CO2 for three days prior to the application of cyclic load stimulation.
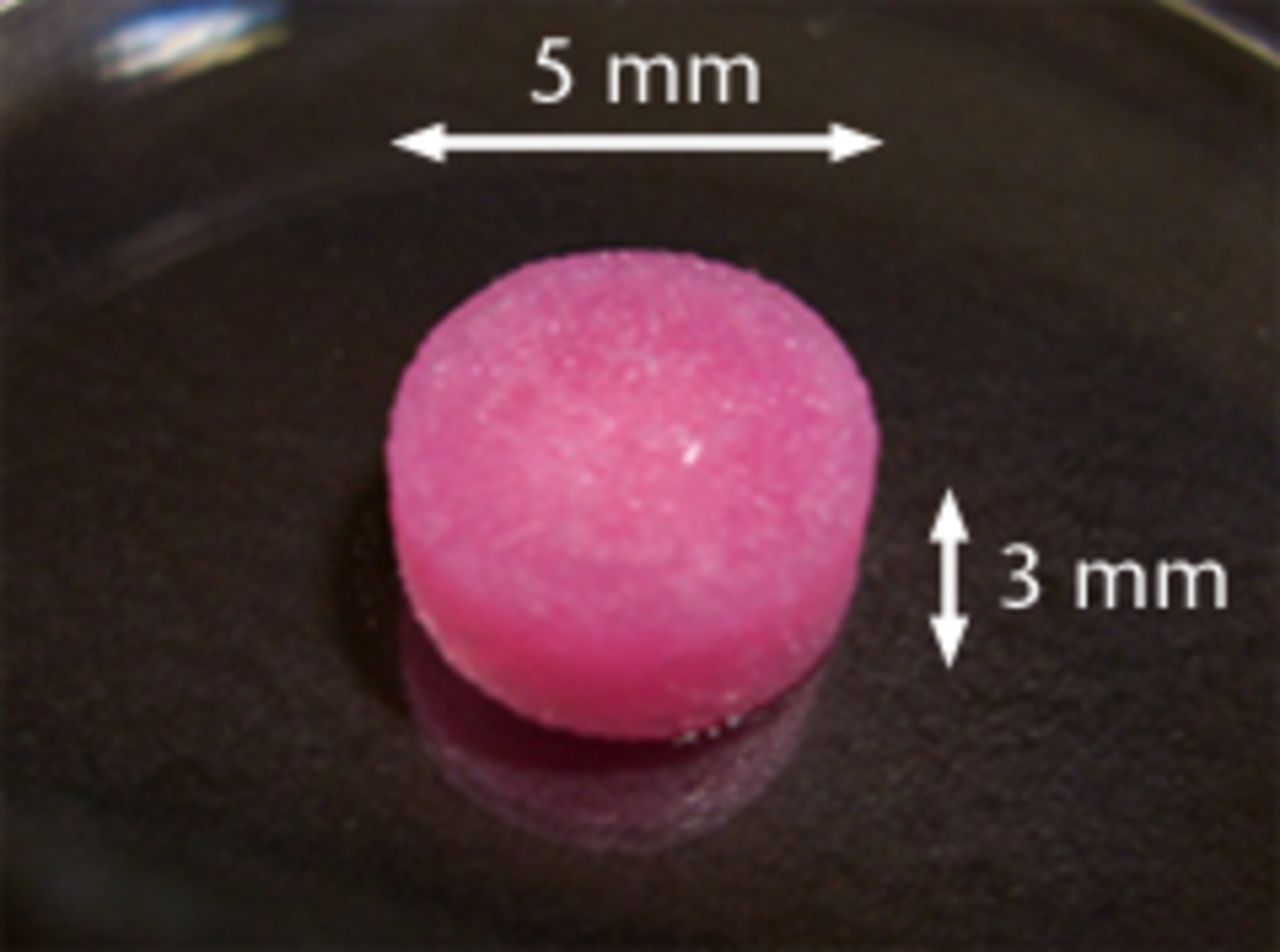


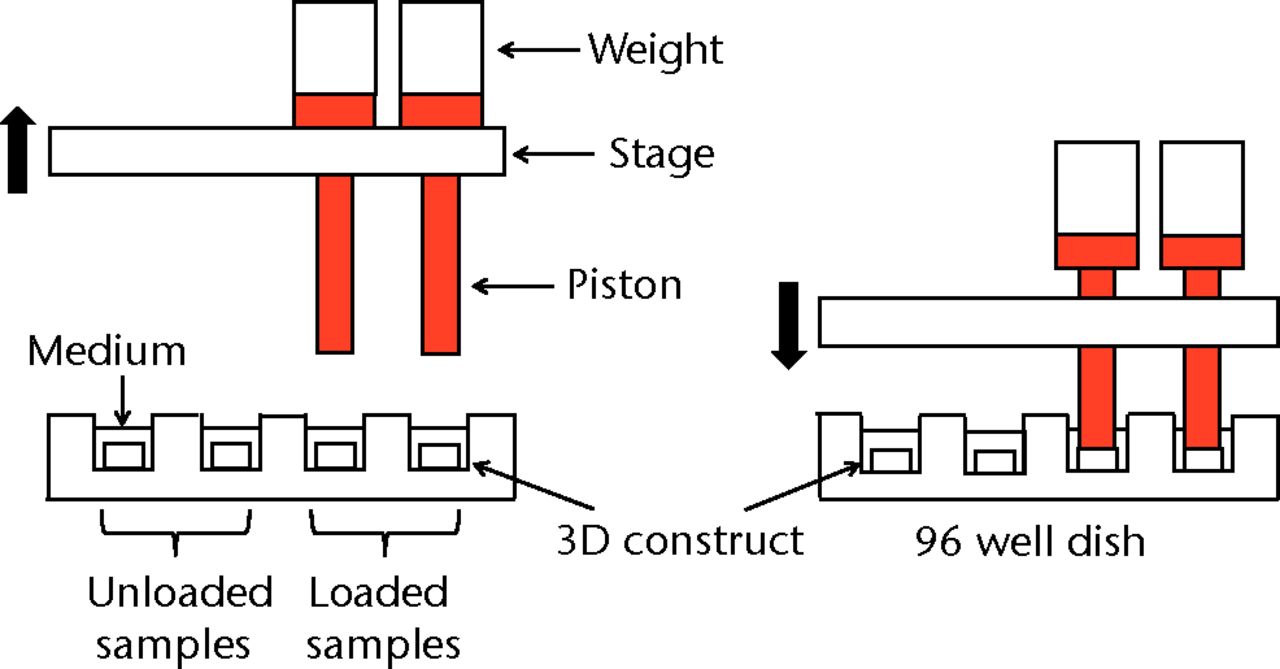
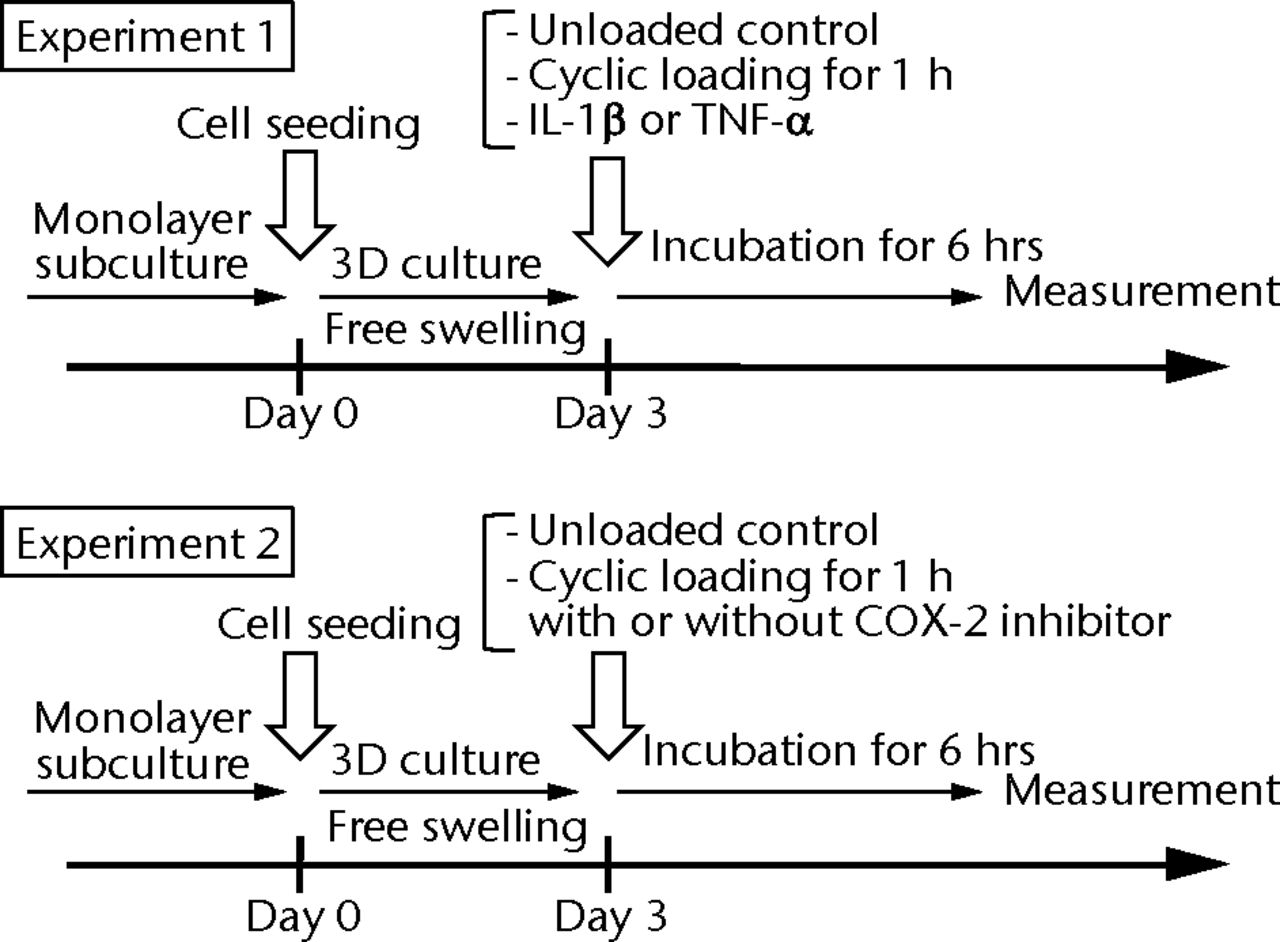
Figs. 1a - 1e
Figure 1a – 3D cell–scaffold constructs made using collagen scaffolds (AtelloCell, MIGHTY); b) monitor and controller; c) Cyclic load stimulater (CLS-5J-Z, Technoview, Osaka, Japan) in the incubator; d) Schematic representation of the cyclic load stimulator, cyclic-loaded samples, and unloaded samples; e) Experimental protocol for cyclic compressive loading on 3D constructs.
Cyclic compressive loading on 3D constructs
Cyclic unconfined compressive loading was applied to the 3D constructs using a custom-designed apparatus, a cyclic load bioreactor (CLS-5J-Z, Technoview, Osaka, Japan), as previously described (Figs 1b to 1d).23 In brief, the loading experiments were performed with metal platens and plastic culture dishes in HG-DMEM and 10% FBS in a humidified incubator maintained at a temperature of 37°C in 5% CO2 . In all of these experiments, a cyclic compressive load of 40 kPa was applied to the constructs for one hour at the rate of 0.5 Hz, in accordance with the protocol used previously, in order to detect the expression of PGE2, IL-1β, IL-6, IL-8 and TNF-α more easily.22,23 As mentioned above, a cyclic compressive load of 40 kPa was chosen, which yielded a 10% compression strain (approximately), because it maximally induced the mRNA expression of MMP1, MMP3, MMP9, MMP13 genes compared with the lower compressive loading of 0 kPa or 20 kPa in our previous study.22 In addition, we measured the expression of PGE2 and the related cytokine expressions six hours after cyclic loading according to our previous study, in which the expression of MMPs maximally upregulated at this time.22
Experimental design
The experimental design is illustrated in Fig. 1e. On day 0, the primary cultured human synovial fibroblasts were harvested, seeded on collagen scaffolds, and maintained in growth media for three days in free-swelling conditions. For the first experiment, on day three, cyclic compressive loading was applied to the 3D constructs for one hour. 3D constructs without loading were considered to be the control. After six hours, culture supernatant was collected, and the concentrations of PGE2, IL-1β, TNF-α, IL-6 and IL-8 were measured with the homogeneous time-resolved fluorescence (HTRF) method (described below in detail). In addition, the mRNA expression of COX-2 and mPGES-1 genes were quantitatively measured using a real-time polymerase chain reaction (PCR). In contrast to the cyclic loading, 10 ng/ml of IL-1β (R& D Systems, Minneapolis, Minnesota) or 100 ng/ml of TNF-α (R& D Systems) was administered to the unloaded 3D constructs on day three. A total of six hours after the administration of these cytokines, the concentration of PGE2 in culture supernatant was measured using HTRF. For the second experiment, cyclic compressive loading was applied to the 3D constructs with or without two types of COX-2 inhibitors: COX-2 selective inhibitor (celecoxib, provided by Pfizer Japan Inc., Tokyo, Japan) or dexamethasone (Sigma-Aldrich). These drugs were administered just before cyclic compressive loading was applied. Six hours after cyclic loading, the concentrations of PGE2, IL-6 and IL-8 in culture supernatant were measured using HTRF. In addition, the mRNA expression of the COX-2 gene was quantitatively estimated by real-time PCR.
Quantitative protein analysis of culture supernatant using HTRF
For each culture supernatant sample, an enzyme immunoassay was performed to measure the concentrations of PGE2, IL-1β, TNF-α, IL-6 and IL-8 using HTRF human PGE2, IL-1β, TNF-α, IL-6 and IL-8 assay kits (CIS Bio International, Saclay, France).
Quantitative mRNA expression analysis of COX-2 and mPGES-1 genes
Total RNAs from the 3D constructs were extracted using a RNeasy mini kit (Qiagen, Valencia, California). Complementary DNAs (cDNAs) were obtained by the use of a reverse transcription (RT) of 200 µg of total RNA through the use of a reverse transcription system (Promega, San Luis Obispo, California) with random primers. For the quantification of gene expression, PCR amplification was performed with SYBR Premix ExTaq (Takara Bio, Shiga, Japan) on a LightCycler 1.5 real-time PCR system (Roche, Indianapolis, Indiana). RNA expression levels were normalised to that of GAPDH. The primers used were as follows: human GAPDH (forward): TCT CTG CTC CTC CTG TTC GAC, (reverse): GTT GAC TCC GAC CTT CAC CTT C, human COX-2 (forward): AGG GTT GCT GGT GGT AGG AA, (reverse): GGT CAA TGG AAG CCT GTG ATA CT, human mPGES-1 (forward): CCT GGG CTT CGT CTA CTC CTT, (reverse): AGT GCA TCC AGG CGA CAA A.
Statistical analysis
Every experiment was performed more than three times using independent donors. Statistical analysis was performed with analysis of variance (ANOVA) followed by post hoc testing (> 2 groups). The comparison of other parameters was analysed with a Mann–Whitney U test (two groups). The results are presented as mean and sd. The data were analysed with JMP 9 (SAS Institute, Cary, North Carolina) and significance was set at p < 0.05.
Results
The expressions of PGE2 and related molecules by cyclic compressive loading
The concentrations of PGE2, IL-6 and IL-8 in a culture supernatant of loaded samples were significantly higher compared with that of unloaded samples (PGE2, 0.33 ng/ml (sd 0.055) vs 2.07 ng/ml (sd 0.65), p < 0.01 (Fig. 2a), p < 0.01 (Fig. 2a); IL-6, 0.71 ng/ml (sd 0.42) vs 6.89 (sd 0.25), p < 0.01 (Fig. 2b), p < 0.01 (Fig. 2b); and IL-8, 0.77 ng/ml (sd 0.39) vs 8.76 ng/ml (sd 0.69), p < 0.01 (Fig. 2c), p < 0.01 (Fig. 2c)). However, the concentrations of IL-1β and TNF-α were unchanged between loaded and unloaded samples (IL-1β, 4.8 pg/ml (sd 8.2) vs 7.4 pg/ml (sd 8.4), p = 0.74 (Fig. 2d), p = 0.74 (Fig. 2d) and TNF-α, 9.6 pg/ml (sd 8.8) vs 7.6 pg/ml (sd 8.3), p = 0.75 (Fig. 2e), p = 0.75 (Fig. 2e)). The administration of IL-1β or TNF-α also significantly induced PGE2 production compared with the non-administered control (IL-1β, 0.33 ng/ml (sd 0.055) vs 2.25 ng/ml (sd 0.65), p < 0.01 and TNF-α, 0.33 ng/ml (sd 0.055) vs 1.84 ng/ml (sd 0.63), p < 0.01 (Fig. 2a), p < 0.01 (Fig. 2a)). The mRNA levels of COX-2 and mPGES-1 genes of loaded samples were significantly higher compared with that of unloaded samples (COX-2, 1 vs 6.97 (sd 3.66), p < 0.01 (Fig. 2f), p < 0.01 (Fig. 2f); mPGES-1, 1 vs 5.03 (sd 2.94), p < 0.01 (Fig. 2g), p < 0.01 (Fig. 2g)).
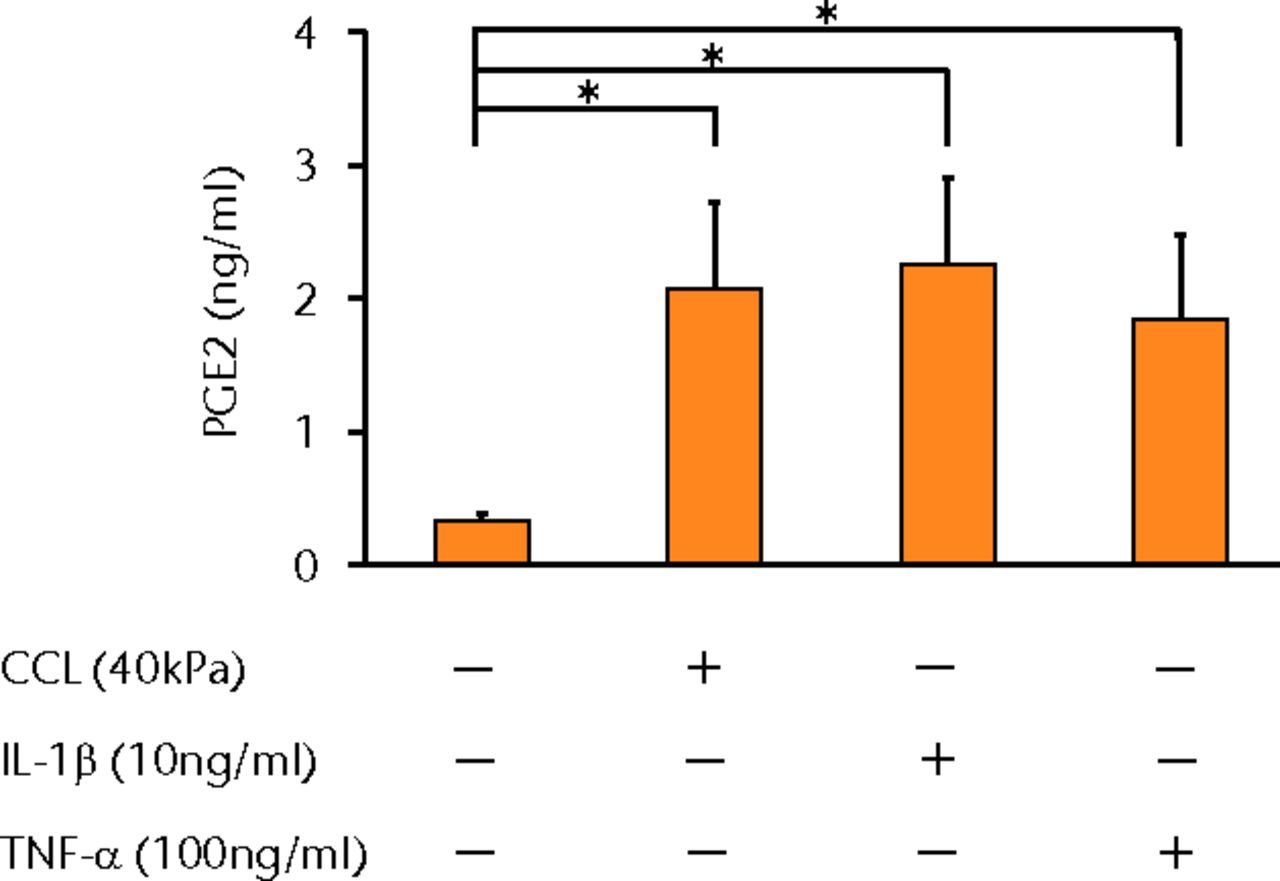
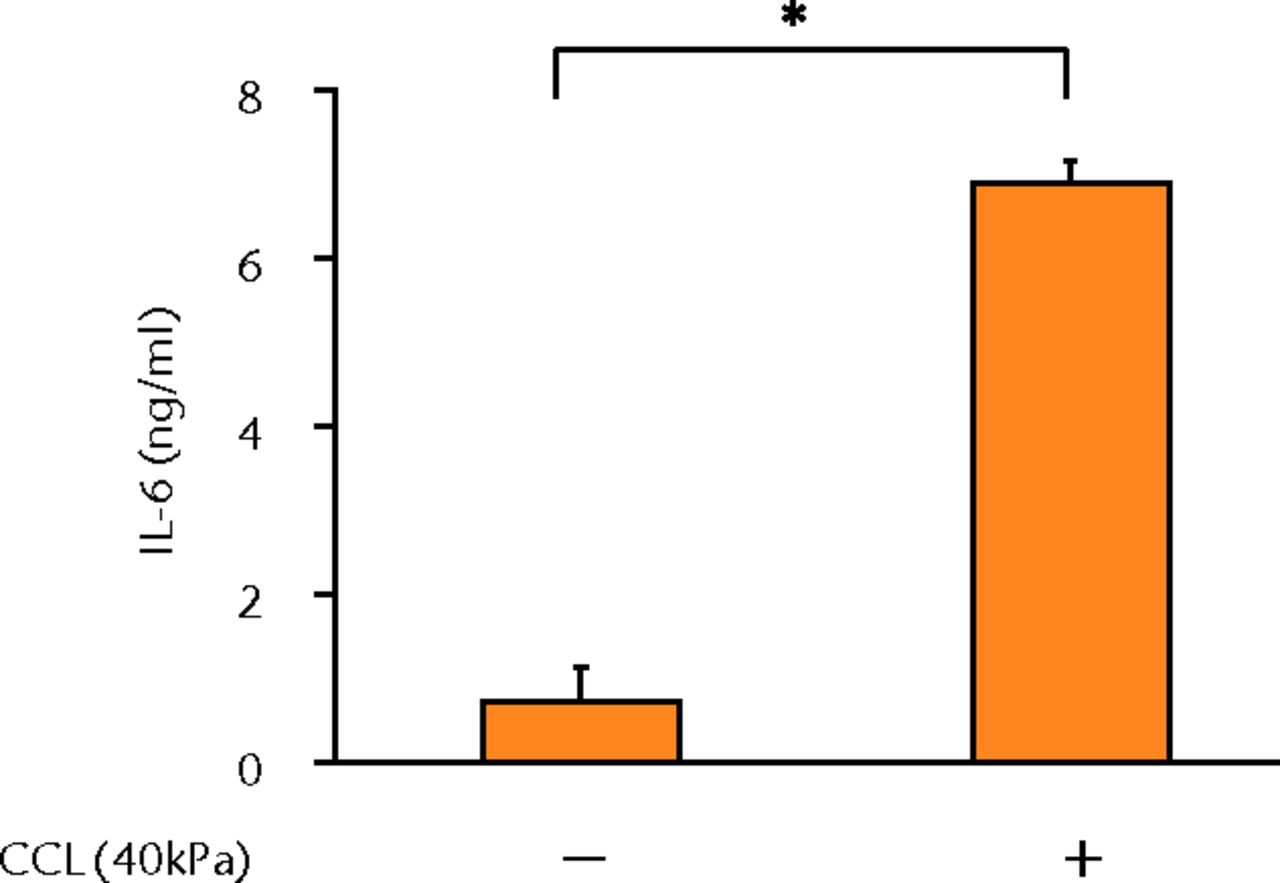
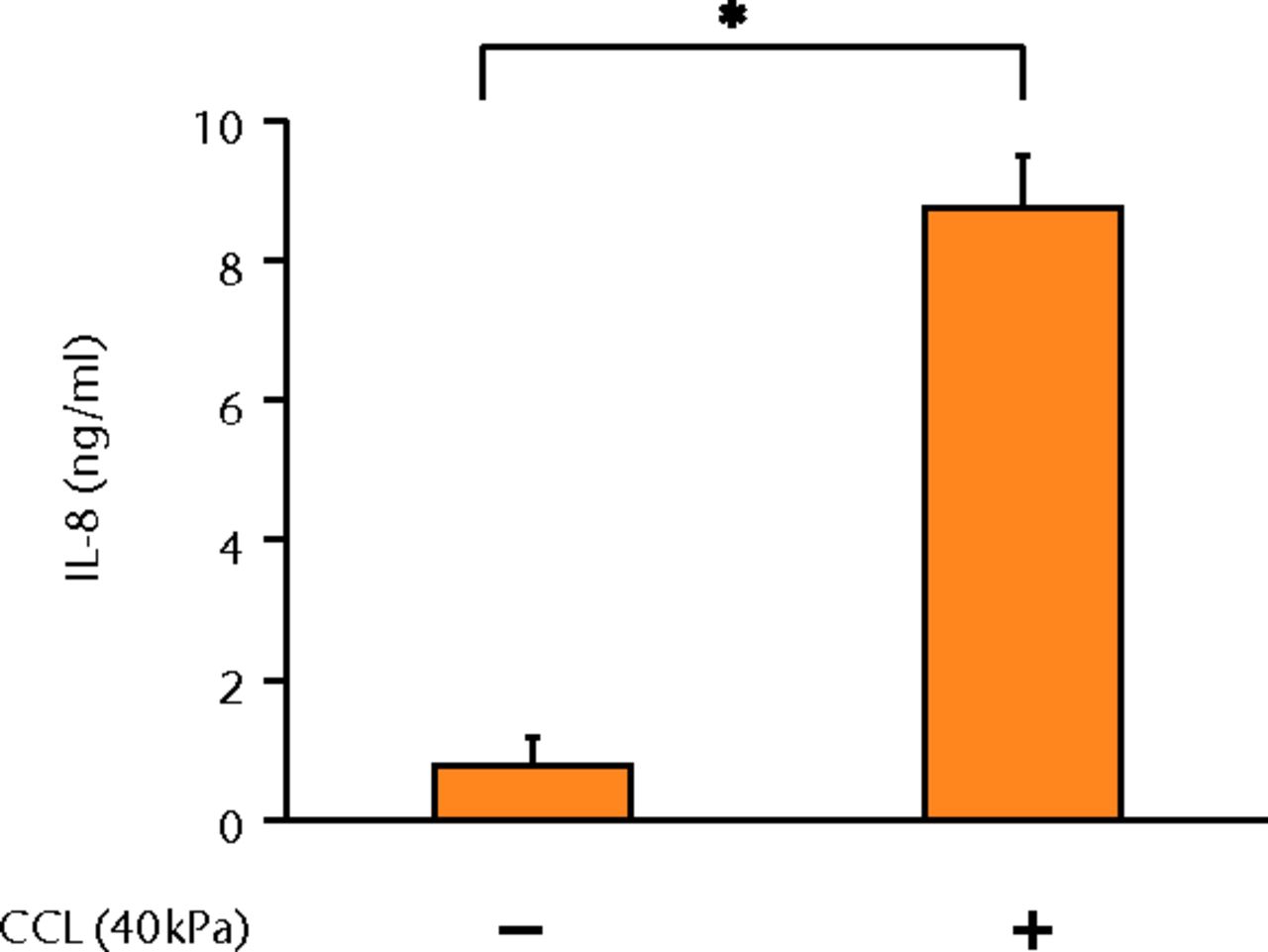
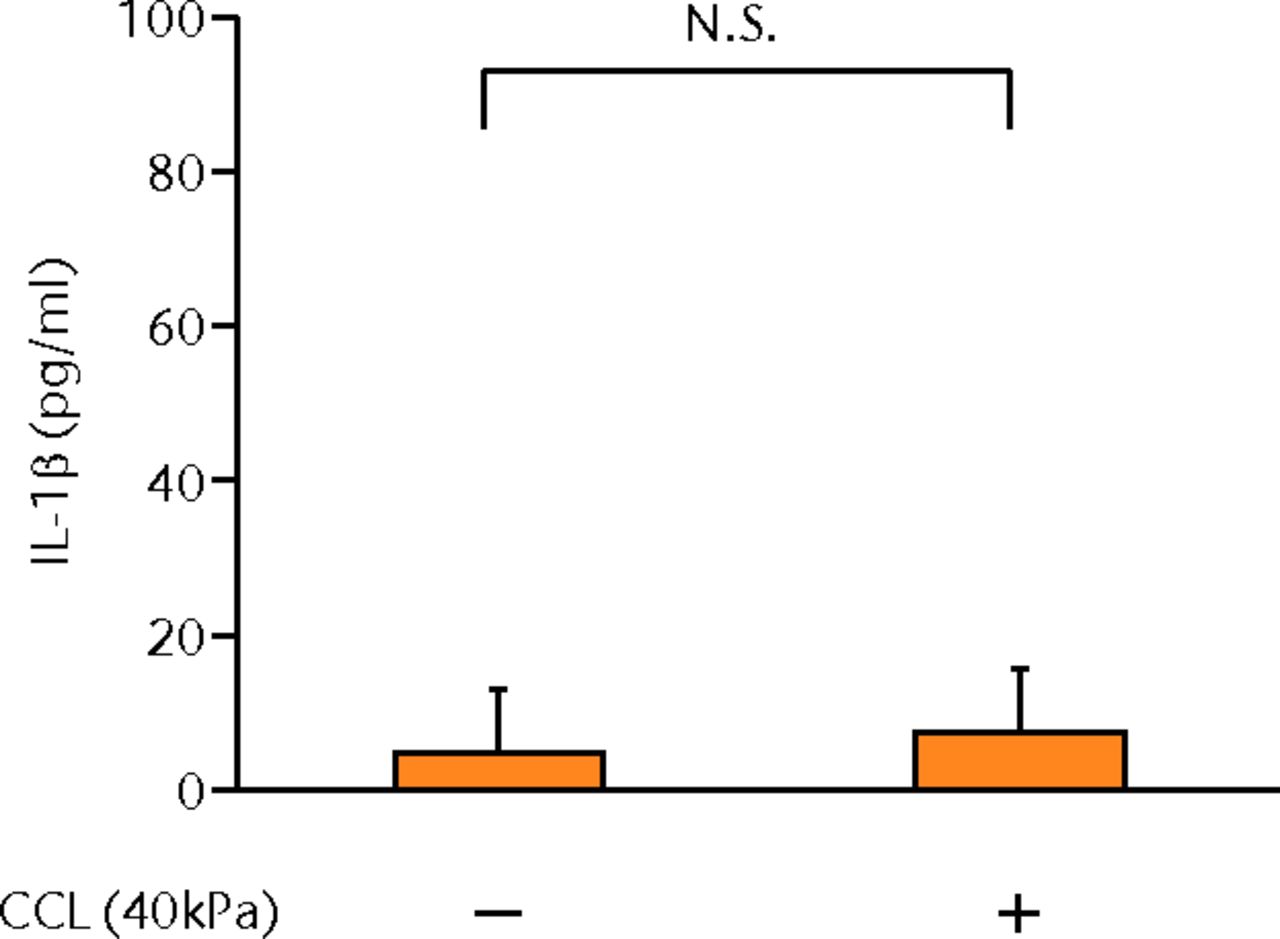
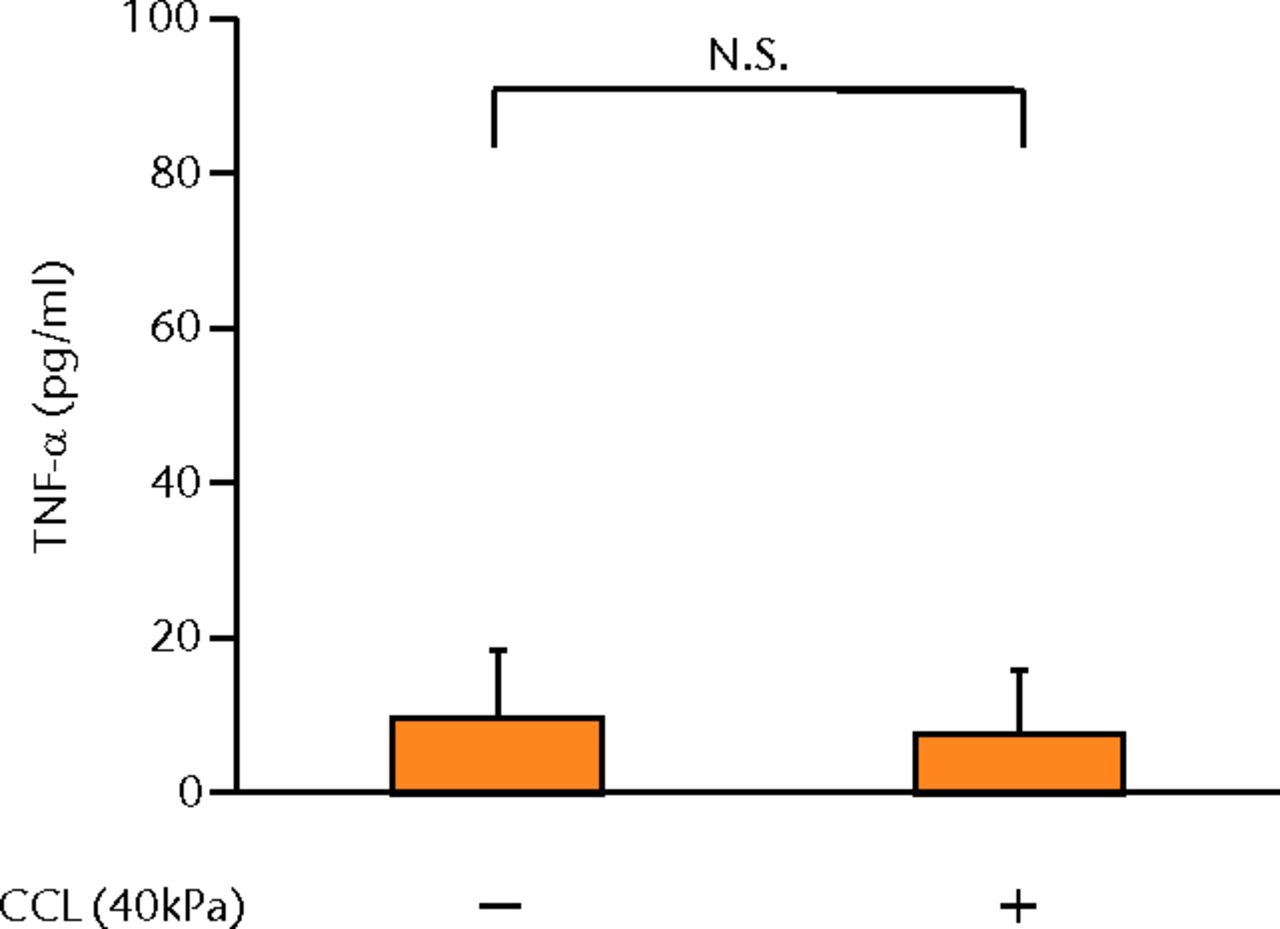
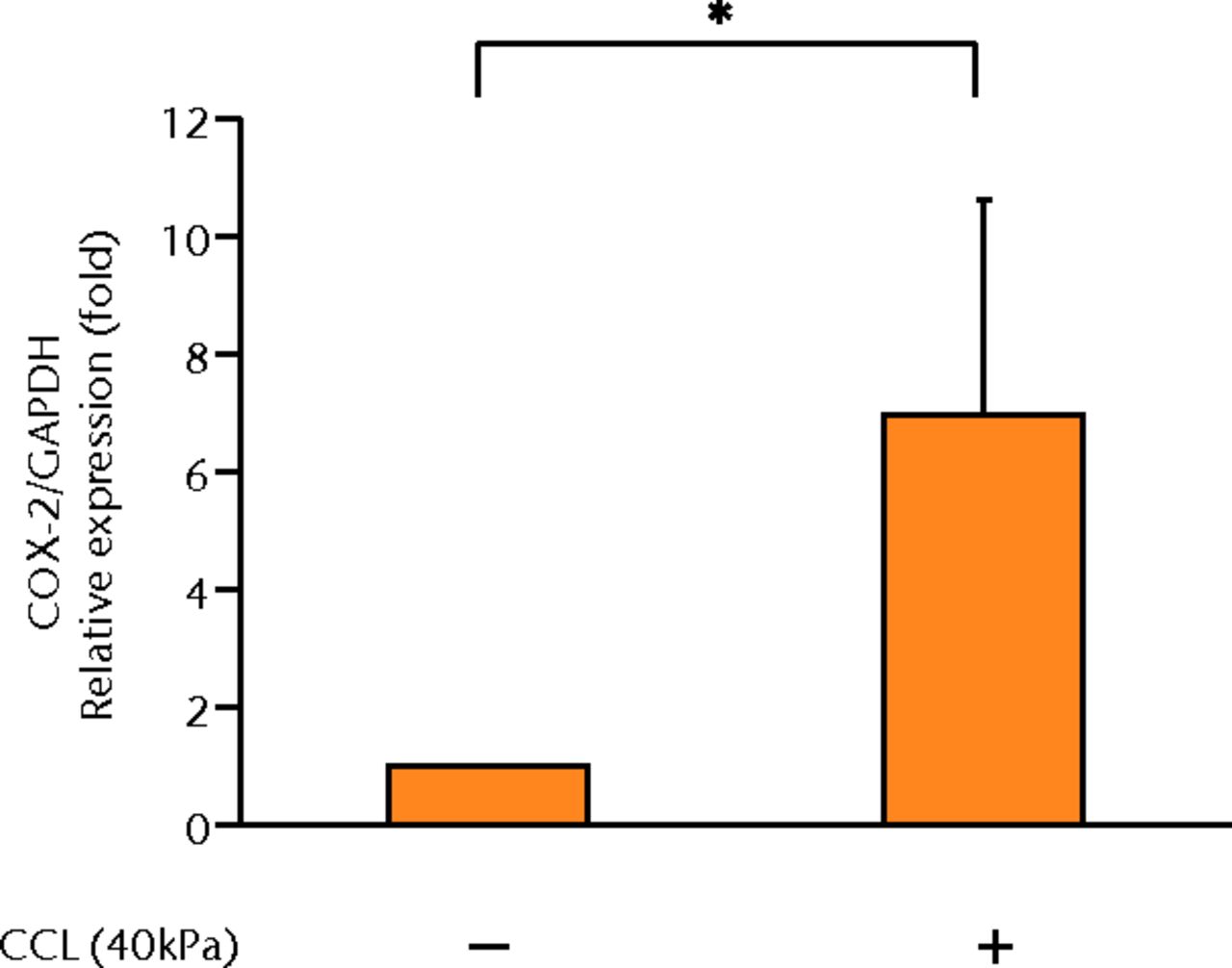
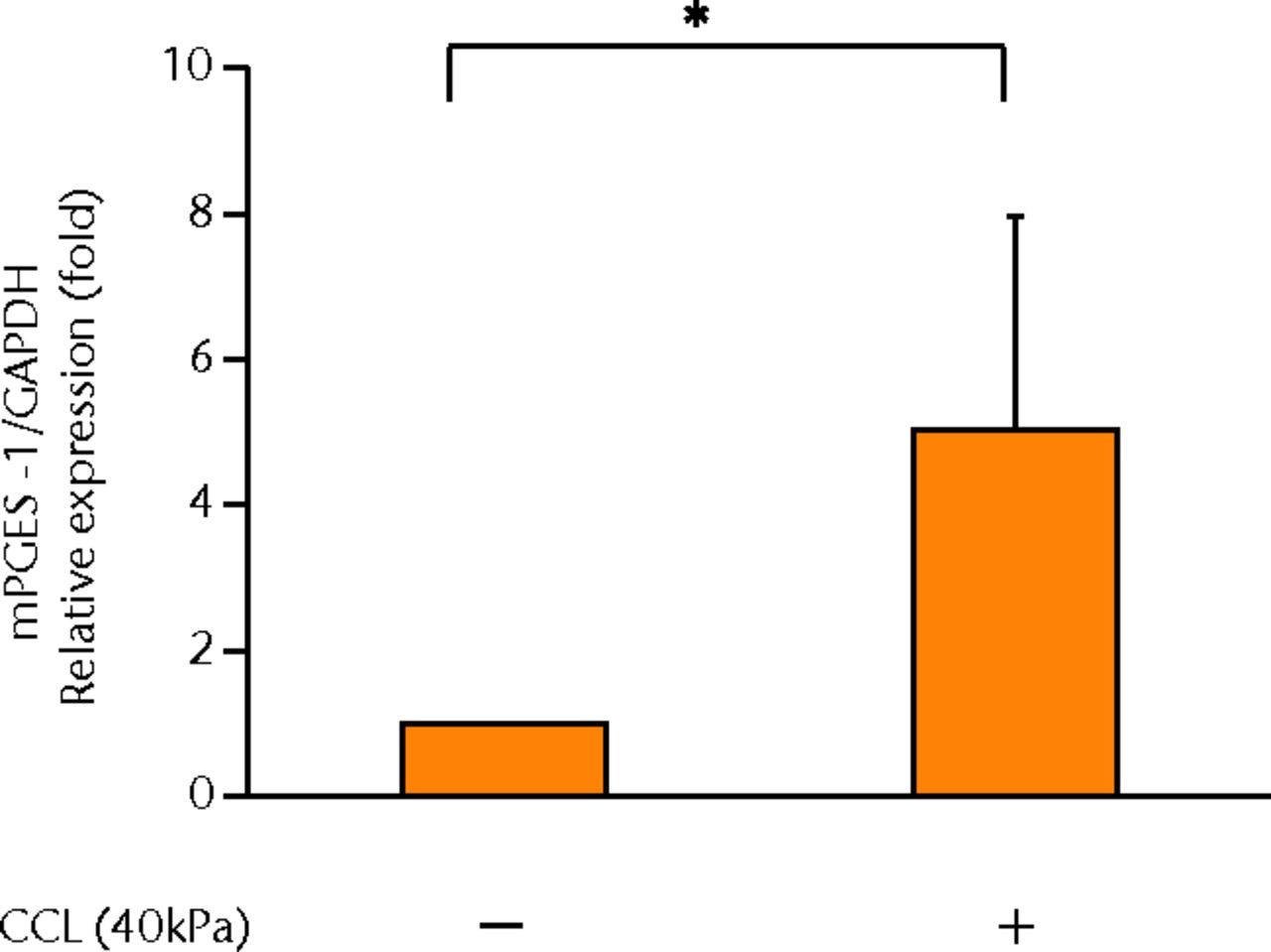
Figs. 2a - 2g
Graphs showing the expressions of PGE2 and related molecules by cyclic compressive loading. a) PGE2 was significantly upregulated by cyclic compressive loading, interleukin- (IL-)1β, or tumour necrosis factor- (TNF)-α (n = 7). b) IL-6 (n = 6) and c) IL-8 (n = 6) were significantly upregulated by cyclic compressive loading. d) IL-1β (n = 6) and e) TNF-α (n = 6) were not upregulated by cyclic compressive loading. f) COX-2 (n = 6) and g) mPGES-1 (n = 5) mRNA levels were significantly upregulated by cyclic compressive loading (CCL;cyclic compressive loading)*p < 0.01.
The effects of a COX-2 selective inhibitor on mechanically induced PGE2, IL-6 and IL-8 proteins and COX-2 gene expressions
The increased concentration of PGE2 by cyclic compressive loading was impeded in a dose-dependent manner after administration of a COX-2 selective inhibitor (Fig. 3a). More than 100 nM of a COX-2 selective inhibitor significantly abolished the upregulation of PGE2 by cyclic compressive loading (p < 0.01). However, the increased concentration of IL-6 and IL-8 by cyclic compressive loading remained high, and the inhibitory effects of the COX-2 selective inhibitor were not observed (Figs 3b and 3c). The upregulation of COX-2 mRNA levels by cyclic compressive loading was not suppressed by a COX-2 selective inhibitor (Fig. 3d). However, the increased concentration of IL-6 and IL-8 by cyclic compressive loading remained high, and the inhibitory effects of the COX-2 selective inhibitor were not observed (Figs 3b and 3c). The upregulation of COX-2 mRNA levels by cyclic compressive loading was not suppressed by a COX-2 selective inhibitor (Fig. 3d). The upregulation of COX-2 mRNA levels by cyclic compressive loading was not suppressed by a COX-2 selective inhibitor (Fig. 3d).
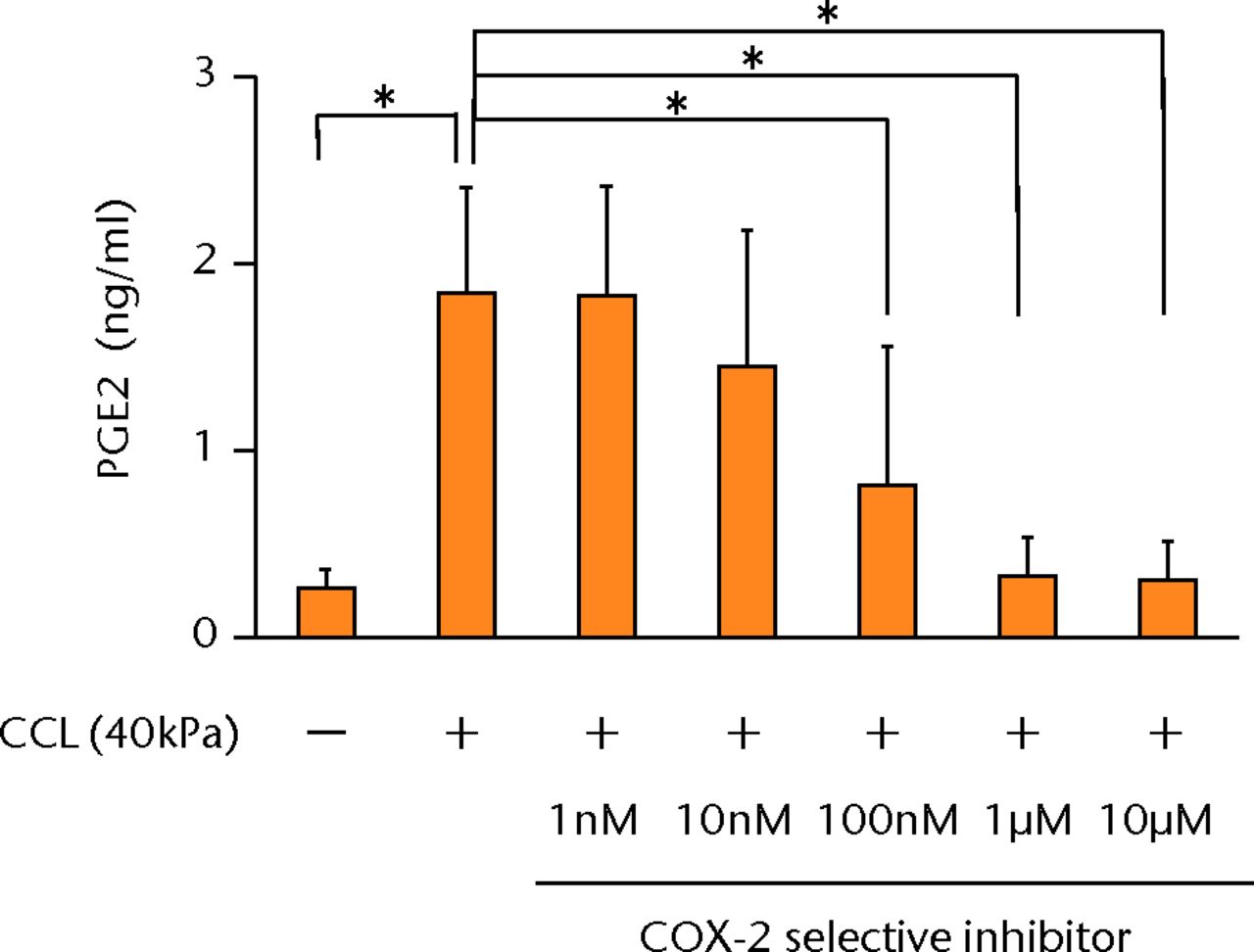
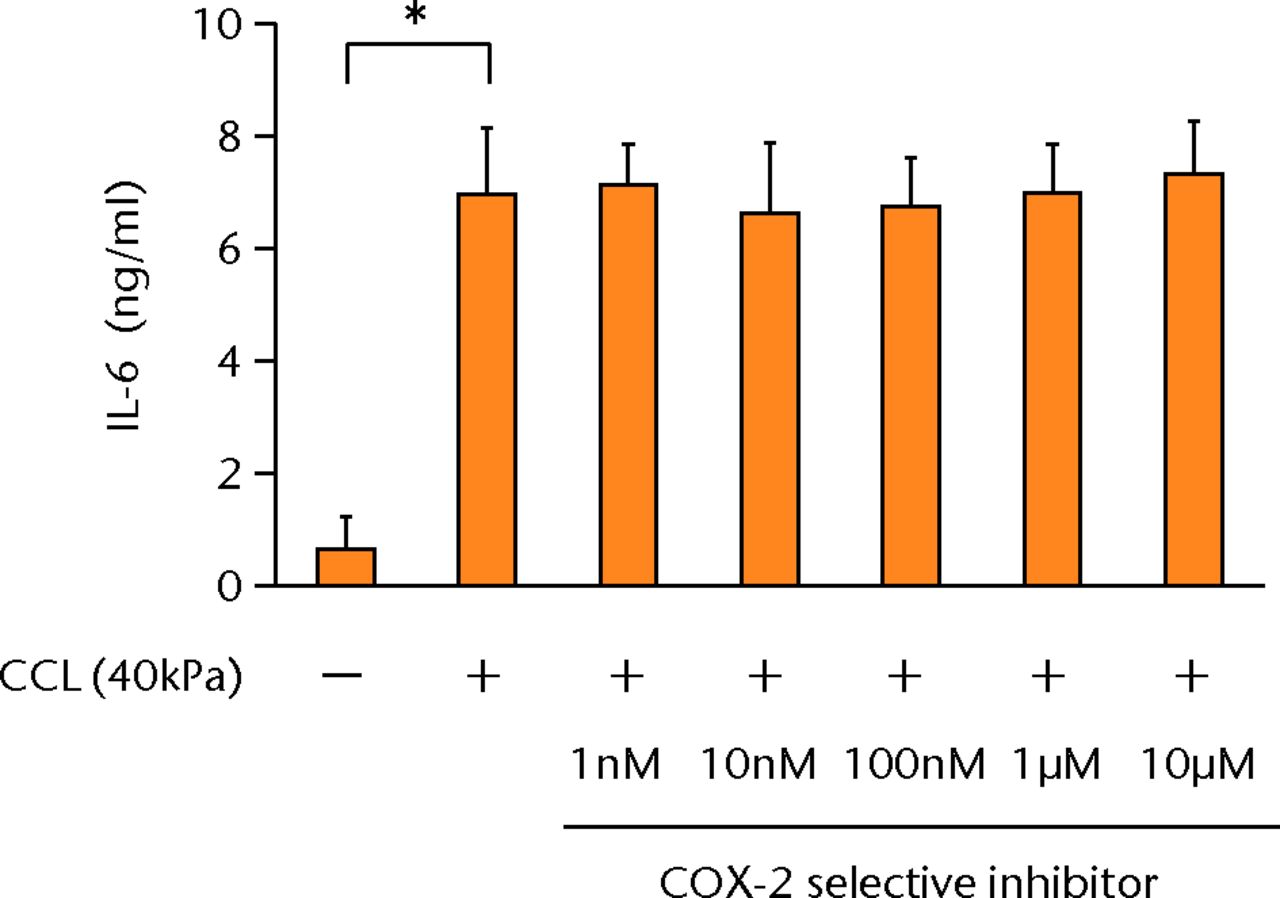
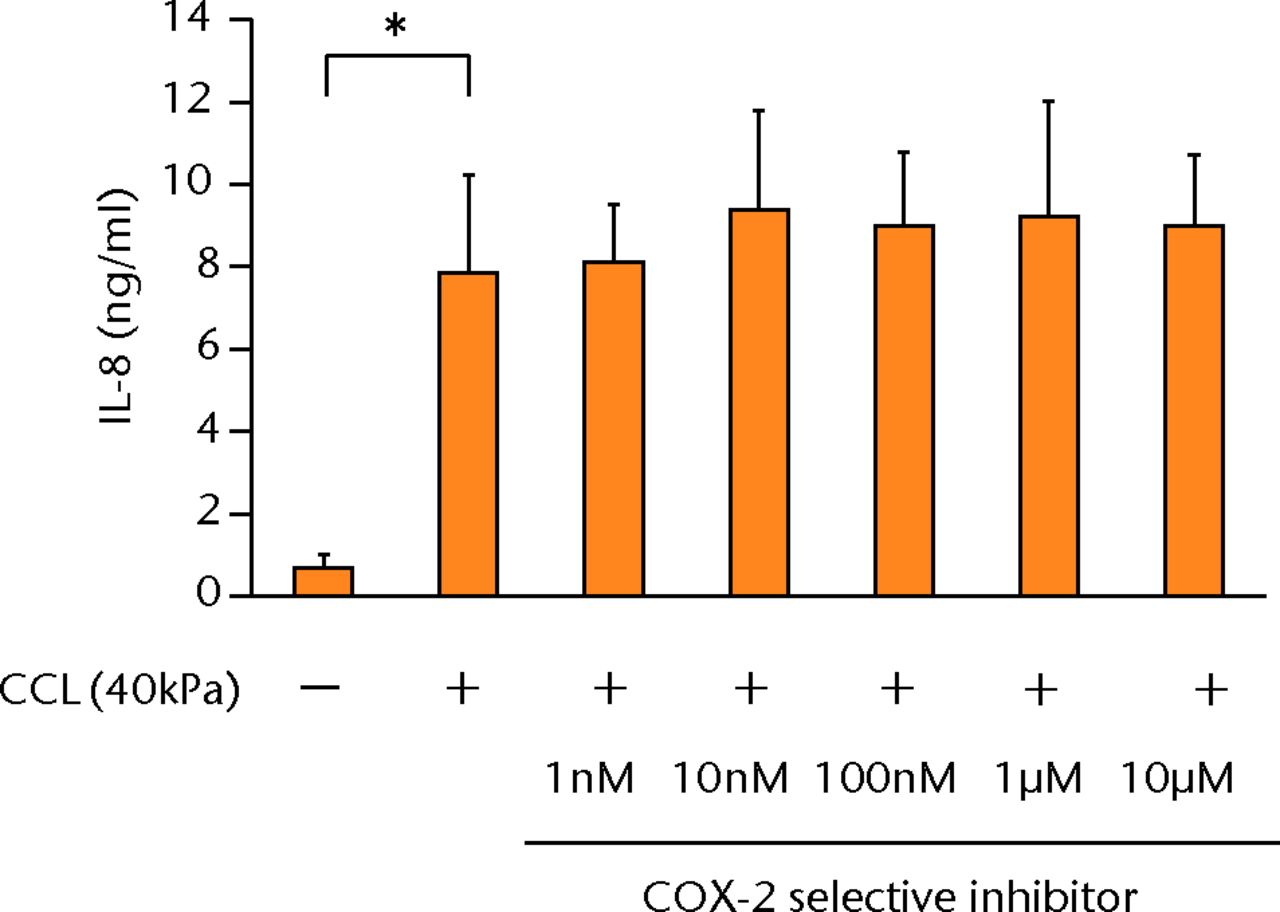
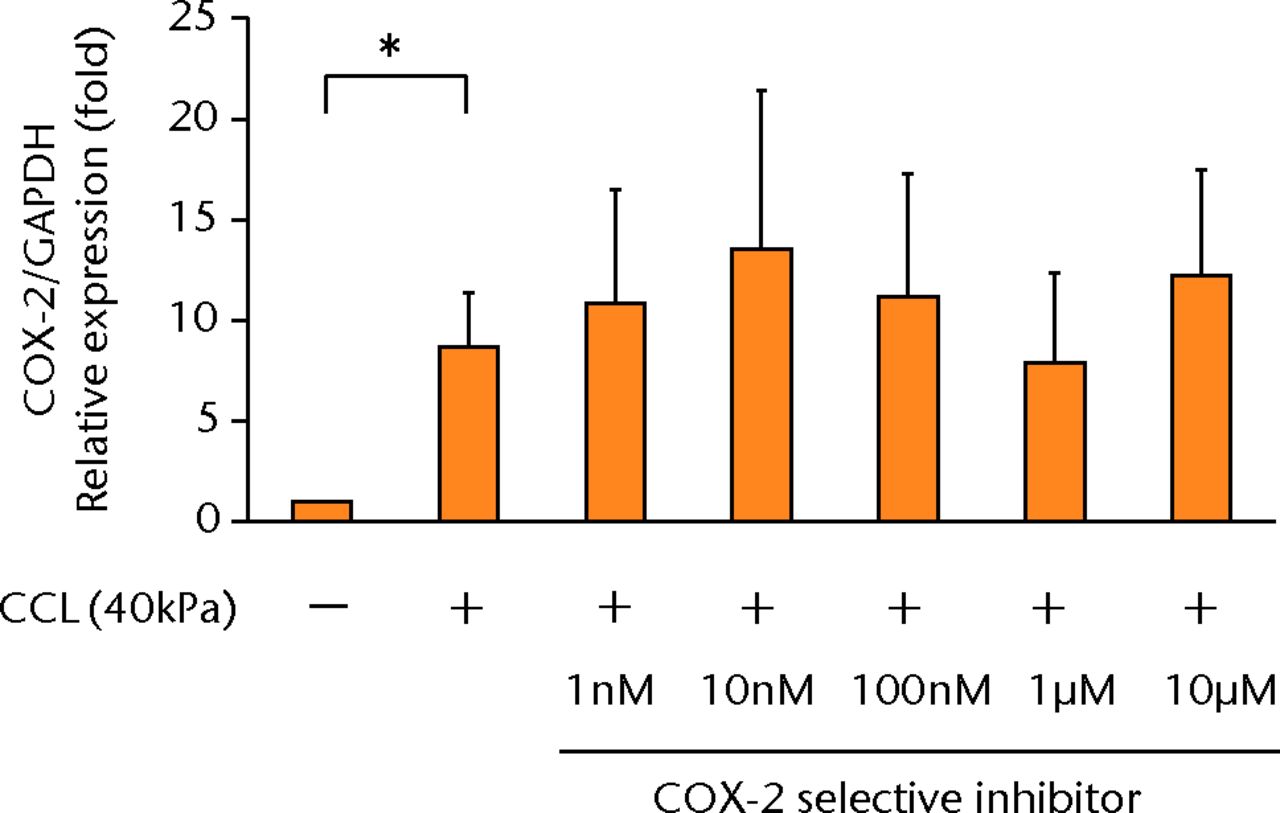
Figs. 3a - 3d
a) The increased concentration of PGE2 by cyclic compressive loading was impeded in a dose-dependent manner (n = 8). b) The increased concentrations of IL-6 (n = 7) and c) IL-8 (n = 5) by cyclic compressive loading remained high. d) The upregulation of COX-2 mRNA levels by cyclic compressive loading was not suppressed by a COX-2 selective inhibitor (n = 5). *p < 0.01
The effects of dexamethasone on mechanically induced PGE2, IL-6 and IL-8 proteins and COX-2 gene expressions
The increased concentration of PGE2 by cyclic compressive loading was suppressed in a dose-dependent manner after administration of dexamethasone (Fig. 4a). More than 100 nM of dexamethasone significantly abolished the upregulation of PGE2 by cyclic compressive loading (p < 0.01). Similarly, the increased concentration of IL-6 and IL-8 was also suppressed in a dose-dependent manner (Figs 4b and 4c). More than 100 nM of dexamethasone significantly abolished the upregulation of IL-6 or IL-8 by cyclic compressive loading (p < 0.01). The upregulation of COX-2 mRNA levels by cyclic compressive loading was suppressed in a dose-dependent manner after the administration of dexamethasone (Fig. 4d). Similarly, the increased concentration of IL-6 and IL-8 was also suppressed in a dose-dependent manner (Figs 4b and 4c). More than 100 nM of dexamethasone significantly abolished the upregulation of IL-6 or IL-8 by cyclic compressive loading (p < 0.01). The upregulation of COX-2 mRNA levels by cyclic compressive loading was suppressed in a dose-dependent manner after the administration of dexamethasone (Fig. 4d). More than 100 nM of dexamethasone significantly abolished the upregulation of IL-6 or IL-8 by cyclic compressive loading (p < 0.01). The upregulation of COX-2 mRNA levels by cyclic compressive loading was suppressed in a dose-dependent manner after the administration of dexamethasone (Fig. 4d). The upregulation of COX-2 mRNA levels by cyclic compressive loading was suppressed in a dose-dependent manner after the administration of dexamethasone (Fig. 4d). More than 10 nM of dexamethasone significantly abolished the upregulation of COX-2 mRNA levels by cyclic compressive loading (p < 0.01).
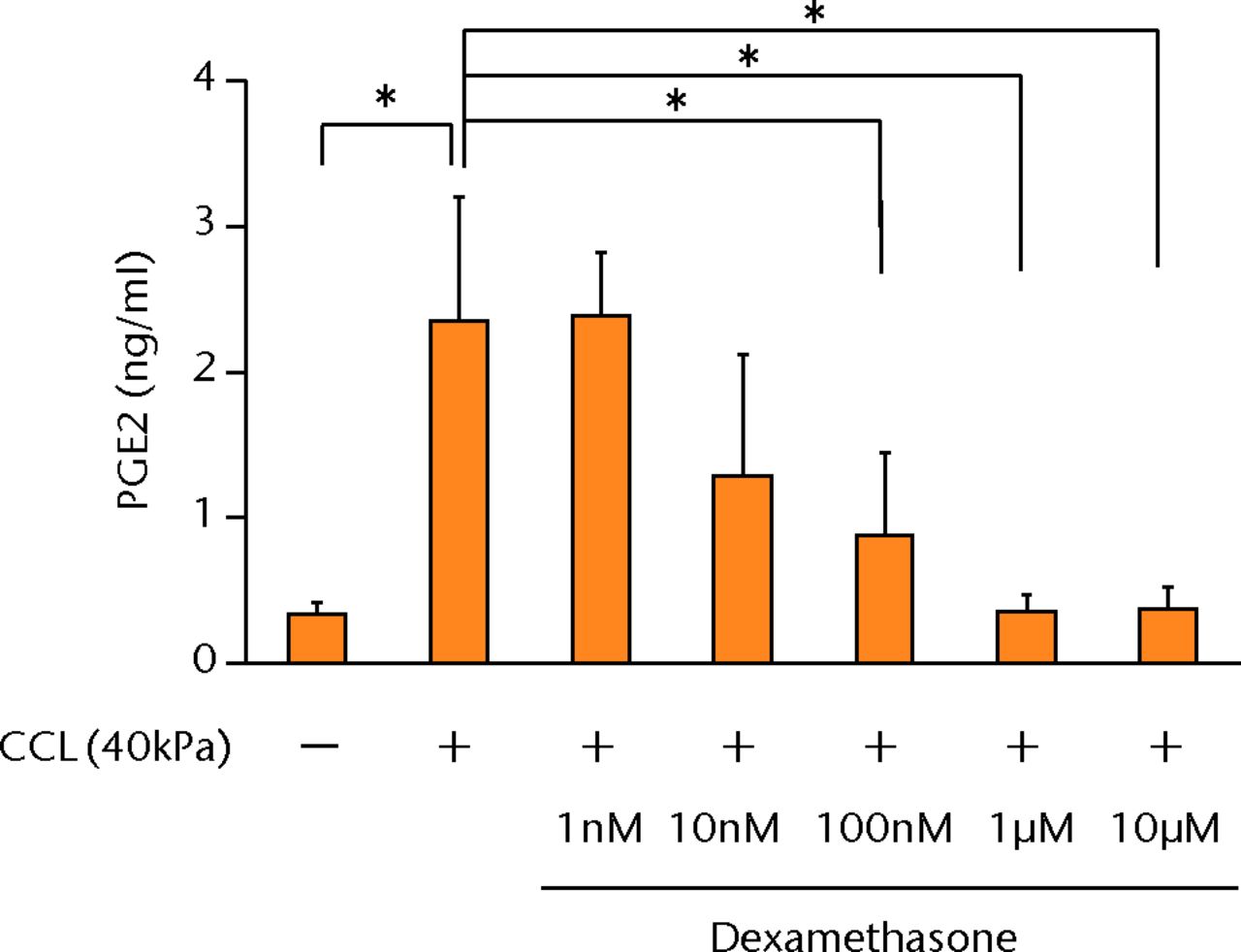
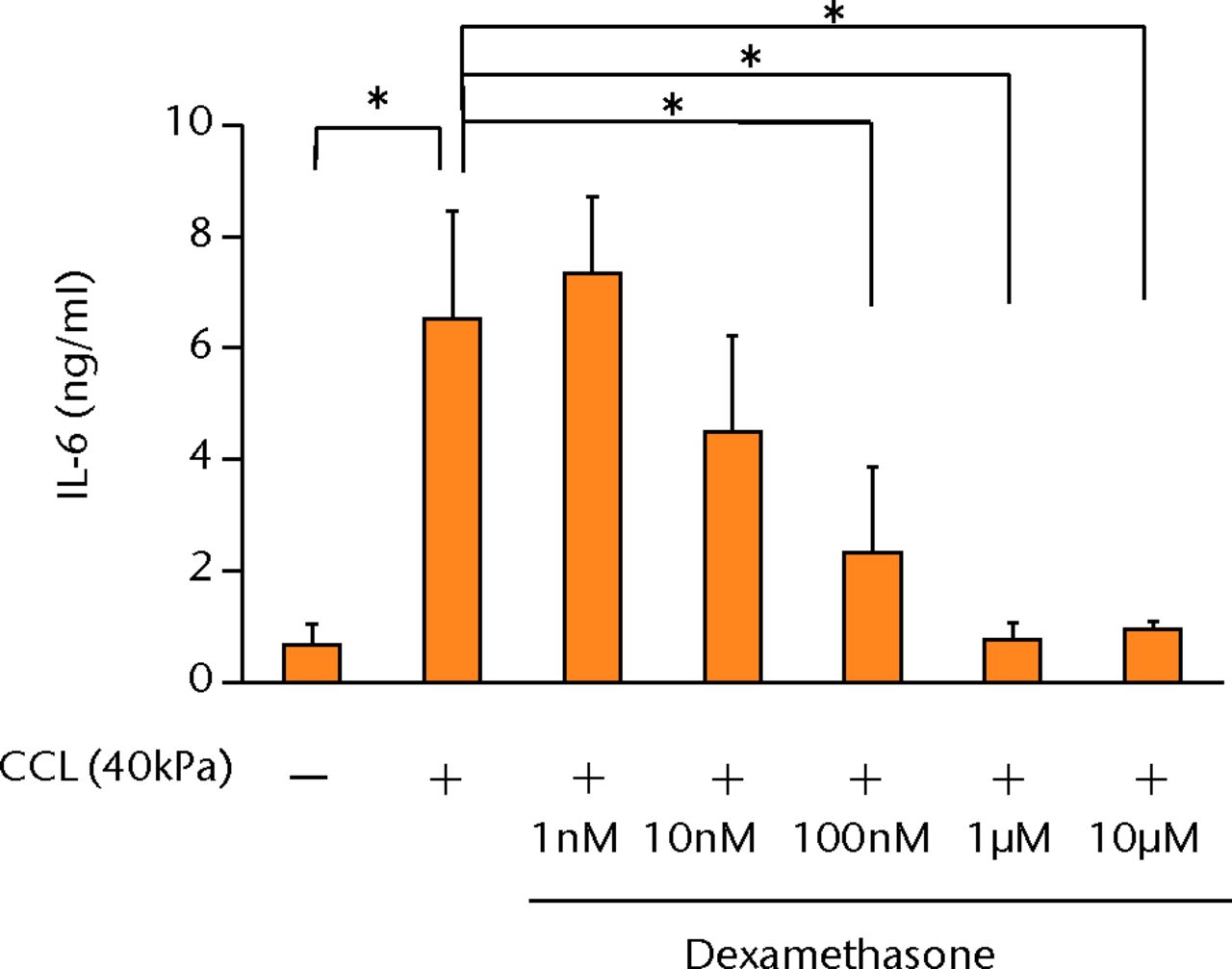
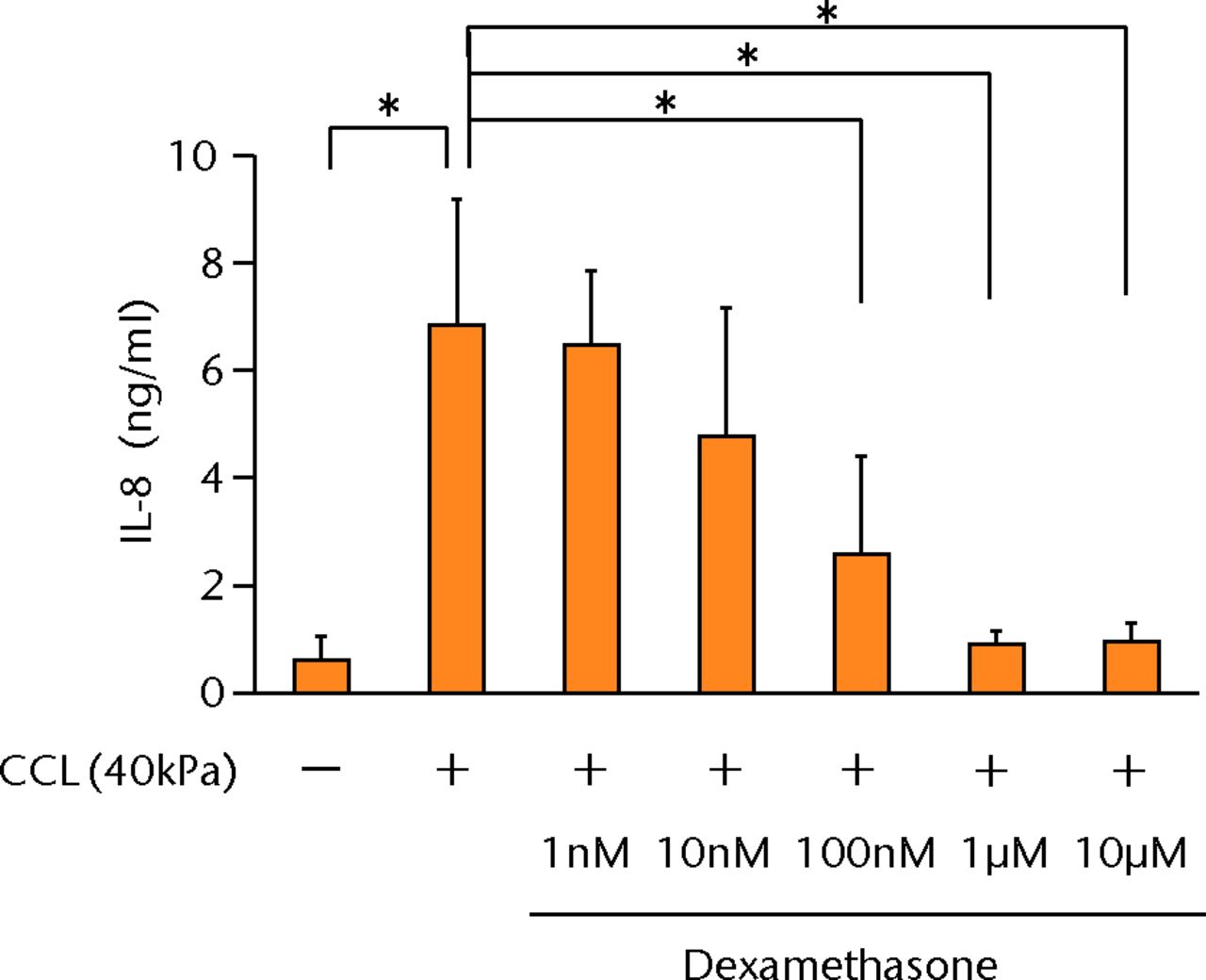
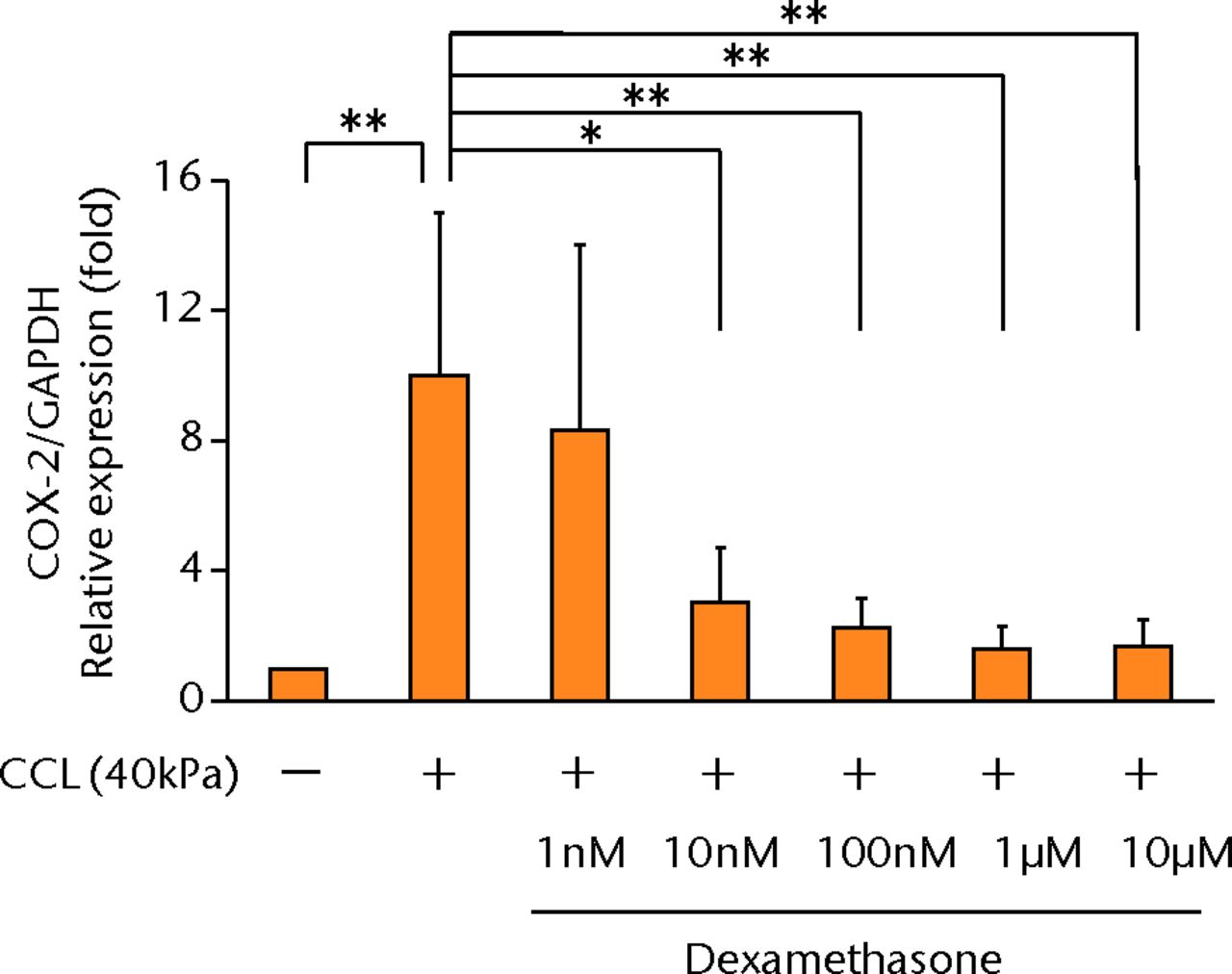
Figs. 4a - 4d
Graphs showing the effects of dexamethasone on mechanically induced PGE2, interleukin- (IL-)6, and IL-8 proteins and COX-2 gene expressions. The increased concentrations of a) PGE2 (n = 5), b) IL-6 (n = 5), and c) IL-8 (n = 5) by cyclic compressive loading were suppressed in a dose-dependent manner. d) The upregulation of COX-2 mRNA levels by cyclic compressive loading was also suppressed in a dose-dependent manner (n = 5). *p < 0.05 **p < 0.01
Discussion
Mechanical stress is believed to be important for every cell in our body, particularly intra-articular tissues such as bone, cartilage, meniscus, and synovium, for the maintenance and regeneration of tissues and organs. Many studies have demonstrated that mechanical stress to chondrocytes, cartilage explants, or mesenchymal stem cells promoted bone and cartilage development.27-30 However, mechanical stress causes joint diseases, and excessive mechanical stress may lead to the development of OA. PGE2 is well known as a pathogenic molecule related to OA development, in addition to MMPs, ADAMTS, and inflammatory cytokines.31 To investigate the molecular mechanisms of PGE2 and related inflammatory cytokines by mechanical stress, we used the 3D culture system using cyclic compressive loading, which can mimic the intra-articular environment through the adjustment of magnitudes, durations, and frequencies of loads. The loading condition of this study represents that cyclic loading for one hour at a rate of 0.5 Hz is nearly equal to the walking pace. We have chosen 40 kPa, which yielded approximately 10% compression strain, because it maximally induced mRNA expression of MMP1, MMP3, MMP9 and MMP13 genes compared with the lower compressive loading of 0 kPa or 20 kPa in our previous study.22 In addition, there have been no obvious data of biomechanics in synovium as far as we know, while > 10% compression strain to cartilage was shown to inhibit proteoglycan and protein synthesis in a dose-dependent manner in bovine calf cartilage.32-34 Therefore, this loading condition may be considered excessive loading over the physiological conditions. Also, the loading was applied with uni-axial unconfined compression, and this condition could also mimic the intra-articular environment, in which both compressive and tensile stresses are applied to the synovium.23,35,36 Moreover, a 3D culture system is better to evaluate a biological reaction, because 3D culture is close to the physical environment and there are sometimes differences detected between 2D and 3D cultures.37-41
In this study, we directly demonstrated that cyclic compressive loading on a 3D-cultured construct of human synovial fibroblasts upregulated PGE2, IL-6 and IL-8 proteins. We also showed that the gene expression of COX-2 and mPGES-1, which are the key enzymes that metabolise arachidonic acid to PGE2, was upregulated by cyclic compressive loading (Fig. 5). In addition, the upregulation of PGE2 by cyclic compressive loading was suppressed by the administrations of a COX-2 selective inhibitor or dexamethasone in a dose-dependent manner. As a pharmacological effect, COX-2 selective inhibitors inhibit the activity of COX-2, whereas dexamethasone inhibits the synthesis of COX-2.42-45 In this study, a COX-2 selective inhibitor suppressed PGE2 production in a dose-dependent manner without changing the COX-2 mRNA level, whereas dexamethasone suppressed PGE2 production by suppressing the COX-2 gene expression. These results reflect well with the pharmacology of PGE2 inhibition by NSAIDs and steroids in OA. Interestingly, a COX-2 selective inhibitor did not suppress IL-6 and IL-8 production, whereas dexamethasone suppressed these cytokines in a dose-dependent manner. The different effects of these chemicals on IL-6 and IL-8 may account for the distinct functions in clinical usage. These functions are still unclear and further studies are required.
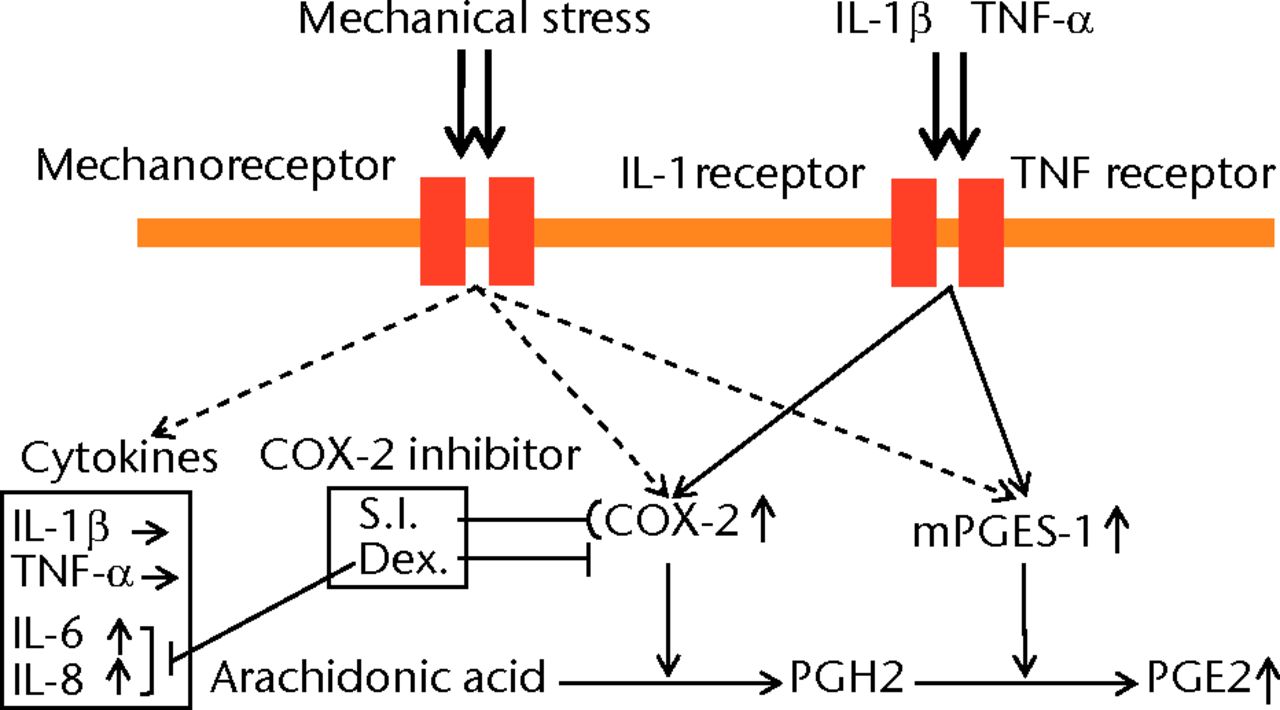
Fig. 5
Schematic representation of the relationship between mechanical stress and the expression of PGE2 and related molecules. Cyclic compressive loading on a 3D cultured construct of human fibroblasts upregulated PGE2, interleukin- (IL-)6 and IL-8 proteins and COX-2, mPGES-1 mRNA levels without IL-1β and tumour necrosis factor- (TNF)-α stimulation. These results indicate that PGE2 upregulation may not be induced via an IL-1β and/or a TNF-α signaling pathway but via other signaling pathways (S.I.; selective inhibitor, Dex; dexamethasone).
Synovial fibroblasts did not produce IL-1β and TNF-α by cyclic compressive loading in this study, as reported previously.6-10 To our surprise, however, synovial fibroblasts produced PGE2, IL-6 and IL-8 without the stimulation of IL-1β and TNF-α, which are produced by synovial macrophages. Undoubtedly, IL-1β and TNF-α, produced by synovial macrophages, are considered to be key factors for OA development through the production of PGE2, MMPs, and ADAMTSs by synovial fibroblasts and chondrocytes.2,5,6,15,20,46-50 On the other hand, it has been unclear what triggers the activation of synovial macrophages. In this study, PGE2 was significantly upregulated by cyclic compressive loading without IL-1β and TNF-α stimulation. Also, we have previously demonstrated that cyclic mechanical stress on synovial fibroblasts upregulated mRNA levels of MMP1, MMP2, MMP3, MMP9, MMP13, ADAMTS4 and ADAMTS5 genes in a load-dependent manner through the same experiment.22,23 Taken together, the upregulation of the key molecules of OA development including PGE2, MMPs, and ADAMTSs was induced by mechanical stress without the upregulation of IL-1β and/or TNF-α. In our opinion, therefore, excessive mechanical stress may ‘switch on’ these gene expressions as the trigger of OA development without IL-1β and/or TNF-α stimulation. (Fig. 5) This notion may coincide with some previous studies using animals and clinical samples, which showed that IL-1β and/or TNF-α were not necessary in OA development.51-53 IL-1β-deficient mice showed development of OA.51 Moreover, the recent clinical study showed that IL-1β and TNF-α in the synovial fluid of patients with OA were not significantly higher than that in the control group.52,53 Therefore, it can be explained that mechanical stress alone is possible to initiate OA development without the stimulation of proinflammatory cytokines.
In a potential limitation of the present study, we did not evaluate other intra-articular cells, such as chondrocytes and meniscal cells. These cells also play an important role in the development of OA. Also, the intracellular signal transductions of PGE2, IL-6 and IL-8 (mechanotransduction) have not been described in detail. In a recent study, mechanotransductions were reported to be related to the Smad pathway,54,55 mitogen-activated protein kinase pathway,56-58 or Wnt signaling pathway.59-61 Our 3D culture system may be useful for the explanation of intracellular mechanotransduction.
In conclusion, cyclic compressive loading on a 3D cultured construct of human fibroblasts upregulated PGE2, IL-6 and IL-8 proteins and COX-2 and mPGES-1 mRNA levels, without IL-1β and TNF-α stimulation. Further investigation may be useful in revealing the molecular mechanism of mechanical stress in vivo for a better understanding of the pathology and therapy of OA.
1 Harris ED Jr . The bone and joint decade: a catalyst for progress. Arthritis Rheum2001;44:1969–1970.CrossrefPubMed Google Scholar
2 Pelletier JP , Martel-PelletierJ, AbramsonSB. Osteoarthritis, an inflammatory disease: potential implication for the selection of new therapeutic targets. Arthritis Rheum2001;44:1237–1247.CrossrefPubMed Google Scholar
3 Farahat MN , YanniG, PostonR, PanayiGS. Cytokine expression in synovial membranes of patients with rheumatoid arthritis and osteoarthritis. Ann Rheum Dis1993;52:870–875.CrossrefPubMed Google Scholar
4 Smith MD , TriantafillouS, ParkerA, YoussefPP, ColemanM. Synovial membrane inflammation and cytokine production in patients with early osteoarthritis. J Rheumatol1997;24:365–371.PubMed Google Scholar
5 Benito MJ , VealeDJ, FitzGeraldO, van den BergWB, BresnihanB. Synovial tissue inflammation in early and late osteoarthritis. Ann Rheum Dis2005;64:1263–1267.CrossrefPubMed Google Scholar
6 Bondeson J , BlomAB, WainwrightS, et al.The role of synovial macrophages and macrophage-produced mediators in driving inflammatory and destructive responses in osteoarthritis. Arthritis Rheum2010;62:647–657.CrossrefPubMed Google Scholar
7 Bartok B , FiresteinGS. Fibroblast-like synoviocytes: key effector cells in rheumatoid arthritis. Immunol Rev2010;233:233–255.CrossrefPubMed Google Scholar
8 Huber LC , DistlerO, TarnerI, et al.Synovial fibroblasts: key players in rheumatoid arthritis. Rheumatology (Oxford)2006;45:669–675.CrossrefPubMed Google Scholar
9 Abeles AM , PillingerMH. The role of the synovial fibroblast in rheumatoid arthritis: cartilage destruction and the regulation of matrix metalloproteinases. Bull NYU Hosp Jt Dis2006;64:20–24.PubMed Google Scholar
10 Smith TJ . Insights into the role of fibroblasts in human autoimmune diseases. Clin Exp Immunol2005;141:388–397.CrossrefPubMed Google Scholar
11 Little CB , MeekerCT, GolubSB, et al.Blocking aggrecanase cleavage in the aggrecan interglobular domain abrogates cartilage erosion and promotes cartilage repair. J Clin Invest2007;117:1627–1636.CrossrefPubMed Google Scholar
12 Glasson SS , AskewR, SheppardB, et al.Deletion of active ADAMTS5 prevents cartilage degradation in a murine model of osteoarthritis. Nature2005;434:644–648.CrossrefPubMed Google Scholar
13 Stanton H , RogersonFM, EastCJ, et al.ADAMTS5 is the major aggrecanase in mouse cartilage in vivo and in vitro. Nature2005;434:648–652.CrossrefPubMed Google Scholar
14 Kawaguchi H . Regulation of osteoarthritis development by Wnt-beta-catenin signaling through the endochondral ossification process. J Bone Miner Res2009;24:8–11.CrossrefPubMed Google Scholar
15 Tortorella MD , MalfaitAM, DeccicoC, ArnerE. The role of ADAM-TS4 (aggrecanase-1) and ADAM-TS5 (aggrecanase-2) in a model of cartilage degradation. Osteoarthritis Cartilage2001;9:539–552.CrossrefPubMed Google Scholar
16 Nishitani K , ItoH, HiramitsuT, et al.PGE2 inhibits MMP expression by suppressing MKK4-JNK MAP kinase-c-JUN pathway via EP4 in human articular chondrocytes. J Cell Biochem2010;109:425–433.CrossrefPubMed Google Scholar
17 Kunisch E , JansenA, KojimaF, et al.Prostaglandin E2 differentially modulates proinflammatory/prodestructive effects of TNF-alpha on synovial fibroblasts via specific E prostanoid receptors/cAMP. J Immunol2009;183:1328–1336.CrossrefPubMed Google Scholar
18 Attur M , Al-MussawirHE, PatelJ, et al.Prostaglandin E2 exerts catabolic effects in osteoarthritis cartilage: evidence for signaling via the EP4 receptor. J Immunol2008;181:5082–5088.CrossrefPubMed Google Scholar
19 Mbalaviele G , PauleyAM, ShafferAF, et al.Distinction of microsomal prostaglandin E synthase-1 (mPGES-1) inhibition from cyclooxygenase-2 inhibition in cells using a novel, selective mPGES-1 inhibitor. Biochem Pharmacol2010;79:1445–1454.CrossrefPubMed Google Scholar
20 Tsutsumi R , ItoH, HiramitsuT, et al.Celecoxib inhibits production of MMP and NO via down-regulation of NF-kappaB and JNK in a PGE2 independent manner in human articular chondrocytes. Rheumatol Int2008;28:727–736.CrossrefPubMed Google Scholar
21 Mastbergen SC , BijlsmaJW, LafeberFP. Selective COX-2 inhibition is favorable to human early and late-stage osteoarthritic cartilage: a human in vitro study. Osteoarthritis Cartilage2005;13:519–526.CrossrefPubMed Google Scholar
22 Akamine Y , KakudoK, KondoM, et al.Prolonged matrix metalloproteinase-3 high expression after cyclic compressive load on human synovial cells in three-dimensional cultured tissue. Int J Oral Maxillofac Surg2012;41:874–881.CrossrefPubMed Google Scholar
23 Muroi Y , KakudoK, NakataK. Effects of compressive loading on human synovium-derived cells. J Dent Res2007;86:786–791.CrossrefPubMed Google Scholar
24 Kojima F , NarabaH, MiyamotoS, et al.Membrane-associated prostaglandin E synthase-1 is upregulated by proinflammatory cytokines in chondrocytes from patients with osteoarthritis. Arthritis Res Ther2004;6:R355–R365.CrossrefPubMed Google Scholar
25 Grimmer C , PfanderD, SwobodaB, et al.Hypoxia-inducible factor 1alpha is involved in the prostaglandin metabolism of osteoarthritic cartilage through up-regulation of microsomal prostaglandin E synthase 1 in articular chondrocytes. Arthritis Rheum2007;56:4084–4094.CrossrefPubMed Google Scholar
26 Ando W , TateishiK, KatakaiD, et al.In vitro generation of a scaffold-free tissue-engineered construct (TEC) derived from human synovial mesenchymal stem cells: biological and mechanical properties and further chondrogenic potential. Tissue Eng Part A2008;14:2041–2049.CrossrefPubMed Google Scholar
27 Takahashi I , NuckollsGH, TakahashiK, et al.Compressive force promotes sox9, type II collagen and aggrecan and inhibits IL-1beta expression resulting in chondrogenesis in mouse embryonic limb bud mesenchymal cells. J Cell Sci1998;111(Pt14):2067–2076.CrossrefPubMed Google Scholar
28 Hunter CJ , MouwJK, LevenstonME. Dynamic compression of chondrocyte-seeded fibrin gels: effects on matrix accumulation and mechanical stiffness. Osteoarthritis Cartilage2004;12:117–130.CrossrefPubMed Google Scholar
29 Fitzgerald JB , JinM, DeanD, et al.Mechanical compression of cartilage explants induces multiple time-dependent gene expression patterns and involves intracellular calcium and cyclic AMP. J Biol Chem2004;279:19502–19511.CrossrefPubMed Google Scholar
30 Vincent TL , HermanssonMA, HansenUN, AmisAA, SaklatvalaJ. Basic fibroblast growth factor mediates transduction of mechanical signals when articular cartilage is loaded. Arthritis Rheum2004;50:526–533.CrossrefPubMed Google Scholar
31 Abramson SB , AtturM. Developments in the scientific understanding of osteoarthritis. Arthritis Res Ther2009;11:227.CrossrefPubMed Google Scholar
32 Sah RL , KimYJ, DoongJY, et al.Biosynthetic response of cartilage explants to dynamic compression. J Orthop Res1989;7:619–636.CrossrefPubMed Google Scholar
33 Sah RL , DoongJY, GrodzinskyAJ, PlaasAH, SandyJD. Effects of compression on the loss of newly synthesized proteoglycans and proteins from cartilage explants. Arch Biochem Biophys1991;286:20–29.CrossrefPubMed Google Scholar
34 Kuettner KE , ColeAA. Cartilage degeneration in different human joints. Osteoarthritis Cartilage2005;13:93–103.CrossrefPubMed Google Scholar
35 Levick JR , McDonaldJN. Ultrastructure of transport pathways in stressed synovium of the knee in anaesthetized rabbits. J Physiol1989;419:493–508.CrossrefPubMed Google Scholar
36 Schett G , Tohidast-AkradM, SteinerG, SmolenJ. The stressed synovium. Arthritis Res2001;3:80–86.CrossrefPubMed Google Scholar
37 Sasazaki Y , SeedhomBB, ShoreR. Morphology of the bovine chondrocyte and of its cytoskeleton in isolation and in situ: are chondrocytes ubiquitously paired through the entire layer of articular cartilage?Rheumatology (Oxford)2008;47:1641–1646.CrossrefPubMed Google Scholar
38 Lin Z , WillersC, XuJ, ZhengMH. The chondrocyte: biology and clinical application. Tissue Eng2006;12:1971–1984.CrossrefPubMed Google Scholar
39 Cukierman E , PankovR, StevensDR, YamadaKM. Taking cell-matrix adhesions to the third dimension. Science2001;294:1708–1712.CrossrefPubMed Google Scholar
40 Schmeichel KL , BissellMJ. Modeling tissue-specific signaling and organ function in three dimensions. J Cell Sci2003;116(Pt12):2377–2388.CrossrefPubMed Google Scholar
41 Hwang NS , KimMS, SampattavanichS, et al.Effects of three-dimensional culture and growth factors on the chondrogenic differentiation of murine embryonic stem cells. Stem Cells2006;24:284–291.CrossrefPubMed Google Scholar
42 Simon LS , LanzaFL, LipskyPE, et al.Preliminary study of the safety and efficacy of SC-58635, a novel cyclooxygenase 2 inhibitor: efficacy and safety in two placebo-controlled trials in osteoarthritis and rheumatoid arthritis, and studies of gastrointestinal and platelet effects. Arthritis Rheum1998;41:1591–1602.CrossrefPubMed Google Scholar
43 Mitchell JA , AkarasereenontP, ThiemermannC, FlowerRJ, VaneJR. Selectivity of nonsteroidal antiinflammatory drugs as inhibitors of constitutive and inducible cyclooxygenase. Proc Natl Acad Sci U S A1993;90:11693–11697.CrossrefPubMed Google Scholar
44 Koyama Y , MizobataT, YamamotoN, et al.Endothelins stimulate expression of cyclooxygenase 2 in rat cultured astrocytes. J Neurochem1999;73:1004–1011.CrossrefPubMed Google Scholar
45 Kujubu DA , HerschmanHR. Dexamethasone inhibits mitogen induction of the TIS10 prostaglandin synthase/cyclooxygenase gene. J Biol Chem1992;267:7991–7994.PubMed Google Scholar
46 Shimpo H , SakaiT, KondoS, et al.Regulation of prostaglandin E(2) synthesis in cells derived from chondrocytes of patients with osteoarthritis. J Orthop Sci2009;14:611–617.CrossrefPubMed Google Scholar
47 Rai MF , RachakondaPS, ManningK, et al.Quantification of cytokines and inflammatory mediators in a three-dimensional model of inflammatory arthritis. Cytokine2008;42:8–17.CrossrefPubMed Google Scholar
48 Scott I , MidhaA, RashidU, et al.Correlation of gene and mediator expression with clinical endpoints in an acute interleukin-1beta-driven model of joint pathology. Osteoarthritis Cartilage2009;17:790–797.CrossrefPubMed Google Scholar
49 Wood DD , IhrieEJ, DinarelloCA, CohenPL. Isolation of an interleukin-1-like factor from human joint effusions. Arthritis Rheum1983;26:975–983.CrossrefPubMed Google Scholar
50 Saha N , MoldovanF, TardifG, et al.Interleukin-1beta-converting enzyme/caspase-1 in human osteoarthritic tissues: localization and role in the maturation of interleukin-1beta and interleukin-18. Arthritis Rheum1999;42:1577–1587.CrossrefPubMed Google Scholar
51 Clements KM , PriceJS, ChambersMG, et al.Gene deletion of either interleukin-1beta, interleukin-1beta-converting enzyme, inducible nitric oxide synthase, or stromelysin 1 accelerates the development of knee osteoarthritis in mice after surgical transection of the medial collateral ligament and partial medial meniscectomy. Arthritis Rheum2003;48:3452–3463.CrossrefPubMed Google Scholar
52 Kokebie R , AggarwalR, LidderS, et al.The role of synovial fluid markers of catabolism and anabolism in osteoarthritis, rheumatoid arthritis and asymptomatic organ donors. Arthritis Res Ther2011;13:R50.CrossrefPubMed Google Scholar
53 Neu CP , ReddiAH, KomvopoulosK, SchmidTM, Di CesarePE. Increased friction coefficient and superficial zone protein expression in patients with advanced osteoarthritis. Arthritis Rheum2010;62:2680–2687.CrossrefPubMed Google Scholar
54 Kido S , Kuriwaka-KidoR, Umino-MiyataniY, et al.Mechanical stress activates Smad pathway through PKCδ to enhance interleukin-11 gene transcription in osteoblasts. PLoS One2010;5:13090.CrossrefPubMed Google Scholar
55 Turner NJ , JonesHS, DaviesJE, CanfieldAE. Cyclic stretch-induced TGFbeta1/Smad signaling inhibits adipogenesis in umbilical cord progenitor cells. Biochem Biophys Res Commun2008;377:1147–1151.CrossrefPubMed Google Scholar
56 Hsu HJ , LeeCF, LockeA, VanderzylSQ, KaunasR. Stretch-induced stress fiber remodeling and the activations of JNK and ERK depend on mechanical strain rate, but not FAK. PLoS One2010;5:12470. Google Scholar
57 Glossop JR , CartmellSH. Effect of fluid flow-induced shear stress on human mesenchymal stem cells: differential gene expression of IL1B and MAP3K8 in MAPK signaling. Gene Expr Patterns2009;9:381–388.CrossrefPubMed Google Scholar
58 Wang BW , ChangH, ShyuKG. Regulation of resistin by cyclic mechanical stretch in cultured rat vascular smooth muscle cells. Clin Sci (Lond)2009;118:221–230.CrossrefPubMed Google Scholar
59 Kitase Y , BarraganL, QingH, et al.Mechanical induction of PGE2 in osteocytes blocks glucocorticoid-induced apoptosis through both the β-catenin and PKA pathways. J Bone Miner Res2010;25:2657–2668.CrossrefPubMed Google Scholar
60 Olkku A , LeskinenJJ, LammiMJ, HynynenK, MahonenA. Ultrasound-induced activation of Wnt signaling in human MG-63 osteoblastic cells. Bone2010;47:320–330.CrossrefPubMed Google Scholar
61 Liedert A , WagnerL, SeefriedL, et al.Estrogen receptor and Wnt signaling interact to regulate early gene expression in response to mechanical strain in osteoblastic cells. Biochem Biophys Res Commun2010;394:755–759.CrossrefPubMed Google Scholar
Funding statement:
This study was supported by a grant from ‘Knowledge Cluster Initiative’ to K. Nakata from The Ministry of Education, Culture, Sports, Science and Technology (MEXT) and Grants-in-Aid for Scientific Research (#23390363 (B)).
Author contributions:
K. Shimomura: Data collection, Data analysis, Writing the paper
T. Kanamoto: Data analysis
K. Kita: Data analysis
Y. Akamine: Data analysis
N. Nakamura: Data analysis
T. Mae: Data analysis
H. Yoshikawa: Data analysis
K. Nakata: Data collection, Data analysis, Writing the paper
ICMJE Conflict of Interest:
None declared
©2014 The British Editorial Society of Bone & Joint Surgery. This is an open-access article distributed under the terms of the Creative Commons Attributions licence, which permits unrestricted use, distribution, and reproduction in any medium, but not for commercial gain, provided the original author and source are credited.










